| 1 |
|
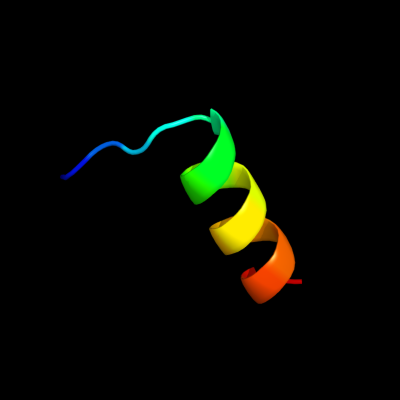
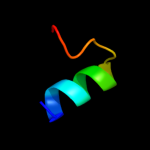
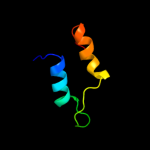
PDB 1m47 chain A
Region: 113 - 129
Aligned: 17
Modelled: 17
Confidence: 42.7%
Identity: 41%
Fold: 4-helical cytokines
Superfamily: 4-helical cytokines
Family: Short-chain cytokines
Phyre2
| 2 |
|
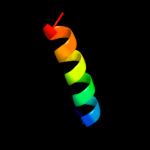
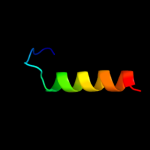
PDB 2keb chain A
Region: 121 - 139
Aligned: 19
Modelled: 19
Confidence: 27.5%
Identity: 5%
PDB header:dna binding protein
Chain: A: PDB Molecule:dna polymerase subunit alpha b;
PDBTitle: nmr solution structure of the n-terminal domain of the dna polymerase2 alpha p68 subunit
Phyre2
| 3 |
|
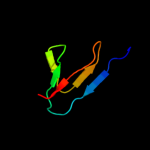
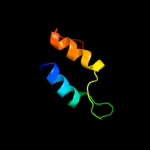
PDB 3ige chain A
Region: 39 - 60
Aligned: 22
Modelled: 22
Confidence: 23.5%
Identity: 41%
PDB header:viral protein
Chain: A: PDB Molecule:soc small outer capsid protein;
PDBTitle: small outer capsid protein (soc) from bacteriophage rb69
Phyre2
| 4 |
|
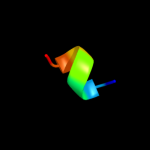
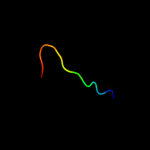
PDB 3b7s chain A domain 1
Region: 103 - 133
Aligned: 31
Modelled: 31
Confidence: 20.3%
Identity: 23%
Fold: alpha-alpha superhelix
Superfamily: ARM repeat
Family: Leukotriene A4 hydrolase C-terminal domain
Phyre2
| 5 |
|
PDB 1f44 chain A domain 1
Region: 122 - 139
Aligned: 18
Modelled: 18
Confidence: 19.3%
Identity: 17%
Fold: SAM domain-like
Superfamily: lambda integrase-like, N-terminal domain
Family: lambda integrase-like, N-terminal domain
Phyre2
| 6 |
|
PDB 3iab chain B
Region: 55 - 81
Aligned: 27
Modelled: 27
Confidence: 15.5%
Identity: 15%
PDB header:hydrolase/rna
Chain: B: PDB Molecule:ribonucleases p/mrp protein subunit pop7;
PDBTitle: crystal structure of rnase p /rnase mrp proteins pop6, pop72 in a complex with the p3 domain of rnase mrp rna
Phyre2
| 7 |
|
PDB 3h6p chain D
Region: 109 - 127
Aligned: 19
Modelled: 19
Confidence: 15.4%
Identity: 32%
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:esat-6-like protein esxr;
PDBTitle: crystal structure of rv3019c-rv3020c from mycobacterium tuberculosis
Phyre2
| 8 |
|
PDB 1iv8 chain A domain 1
Region: 41 - 80
Aligned: 40
Modelled: 40
Confidence: 14.6%
Identity: 28%
Fold: Glycosyl hydrolase domain
Superfamily: Glycosyl hydrolase domain
Family: alpha-Amylases, C-terminal beta-sheet domain
Phyre2
| 9 |
|
PDB 3g36 chain D
Region: 120 - 126
Aligned: 7
Modelled: 7
Confidence: 14.6%
Identity: 57%
PDB header:nuclear protein
Chain: D: PDB Molecule:protein dpy-30 homolog;
PDBTitle: crystal structure of the human dpy-30-like c-terminal domain
Phyre2
| 10 |
|
PDB 2dsy chain A domain 1
Region: 113 - 140
Aligned: 27
Modelled: 28
Confidence: 13.6%
Identity: 26%
Fold: TTHA1013/TTHA0281-like
Superfamily: TTHA1013/TTHA0281-like
Family: TTHA0281-like
Phyre2
| 11 |
|
PDB 3iu0 chain A
Region: 87 - 131
Aligned: 39
Modelled: 45
Confidence: 12.4%
Identity: 26%
PDB header:transferase
Chain: A: PDB Molecule:protein-glutamine gamma-glutamyltransferase;
PDBTitle: structural basis for zymogen activation and substrate binding of2 transglutaminase from streptomyces mobaraense
Phyre2
| 12 |
|
PDB 1xsq chain A
Region: 36 - 61
Aligned: 26
Modelled: 26
Confidence: 10.4%
Identity: 23%
Fold: Double-stranded beta-helix
Superfamily: RmlC-like cupins
Family: Ureidoglycolate hydrolase AllA
Phyre2
| 13 |
|
PDB 1ufo chain A
Region: 104 - 127
Aligned: 24
Modelled: 24
Confidence: 10.1%
Identity: 17%
Fold: alpha/beta-Hydrolases
Superfamily: alpha/beta-Hydrolases
Family: Hypothetical protein TT1662
Phyre2
| 14 |
|
PDB 1mky chain A domain 3
Region: 117 - 126
Aligned: 10
Modelled: 10
Confidence: 9.1%
Identity: 40%
Fold: Alpha-lytic protease prodomain-like
Superfamily: Probable GTPase Der, C-terminal domain
Family: Probable GTPase Der, C-terminal domain
Phyre2
| 15 |
|
PDB 1t15 chain A domain 1
Region: 124 - 134
Aligned: 11
Modelled: 11
Confidence: 8.6%
Identity: 9%
Fold: BRCT domain
Superfamily: BRCT domain
Family: BRCT domain
Phyre2
| 16 |
|
PDB 1xhm chain B domain 1
Region: 123 - 136
Aligned: 14
Modelled: 14
Confidence: 8.4%
Identity: 43%
Fold: Non-globular all-alpha subunits of globular proteins
Superfamily: Transducin (heterotrimeric G protein), gamma chain
Family: Transducin (heterotrimeric G protein), gamma chain
Phyre2
| 17 |
|
PDB 1xhm chain B
Region: 123 - 136
Aligned: 14
Modelled: 14
Confidence: 8.2%
Identity: 43%
PDB header:signaling protein
Chain: B: PDB Molecule:guanine nucleotide-binding protein g(i)/g(s)
PDBTitle: the crystal structure of a biologically active peptide2 (sigk) bound to a g protein beta:gamma heterodimer
Phyre2
| 18 |
|
PDB 1iu4 chain A
Region: 87 - 131
Aligned: 39
Modelled: 45
Confidence: 7.7%
Identity: 26%
Fold: Cysteine proteinases
Superfamily: Cysteine proteinases
Family: Microbial transglutaminase
Phyre2
| 19 |
|
PDB 2jya chain A
Region: 76 - 87
Aligned: 12
Modelled: 12
Confidence: 6.5%
Identity: 33%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein atu1810;
PDBTitle: nmr solution structure of protein atu1810 from agrobacterium2 tumefaciens. northeast structural genomics consortium target atr23,3 ontario centre for structural proteomics target atc1776
Phyre2
| 20 |
|
PDB 1l0b chain A domain 1
Region: 124 - 134
Aligned: 11
Modelled: 11
Confidence: 6.4%
Identity: 9%
Fold: BRCT domain
Superfamily: BRCT domain
Family: BRCT domain
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|