1 c3rgwS_
100.0
76
PDB header: oxidoreductase/oxidoreductaseChain: S: PDB Molecule: membrane-bound hydrogenase (nife) small subunit hoxk;PDBTitle: crystal structure at 1.5 a resolution of an h2-reduced, o2-tolerant2 hydrogenase from ralstonia eutropha unmasks a novel iron-sulfur3 cluster
2 c3myrE_
100.0
47
PDB header: oxidoreductaseChain: E: PDB Molecule: hydrogenase (nife) small subunit hyda;PDBTitle: crystal structure of [nife] hydrogenase from allochromatium vinosum in2 its ni-a state
3 d1wuis1
100.0
41
Fold: HydA/Nqo6-likeSuperfamily: HydA/Nqo6-likeFamily: Nickel-iron hydrogenase, small subunit4 c1h2aS_
100.0
41
PDB header: oxidoreductaseChain: S: PDB Molecule: hydrogenase;PDBTitle: single crystals of hydrogenase from desulfovibrio vulgaris
5 d1frfs_
100.0
45
Fold: HydA/Nqo6-likeSuperfamily: HydA/Nqo6-likeFamily: Nickel-iron hydrogenase, small subunit6 d1e3da_
100.0
42
Fold: HydA/Nqo6-likeSuperfamily: HydA/Nqo6-likeFamily: Nickel-iron hydrogenase, small subunit7 d1yq9a1
100.0
42
Fold: HydA/Nqo6-likeSuperfamily: HydA/Nqo6-likeFamily: Nickel-iron hydrogenase, small subunit8 d1cc1s_
100.0
36
Fold: HydA/Nqo6-likeSuperfamily: HydA/Nqo6-likeFamily: Nickel-iron hydrogenase, small subunit9 c2wpnA_
100.0
34
PDB header: oxidoreductaseChain: A: PDB Molecule: periplasmic [nifese] hydrogenase, small subunit;PDBTitle: structure of the oxidised, as-isolated nifese hydrogenase2 from d. vulgaris hildenborough
10 d2fug61
99.6
23
Fold: HydA/Nqo6-likeSuperfamily: HydA/Nqo6-likeFamily: Nq06-like11 c2g4rB_
89.6
11
PDB header: biosynthetic proteinChain: B: PDB Molecule: molybdopterin biosynthesis mog protein;PDBTitle: anomalous substructure of moga
12 d2ftsa3
88.7
17
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MoeA central domain-like13 c2is8A_
86.7
12
PDB header: structural proteinChain: A: PDB Molecule: molybdopterin biosynthesis enzyme, moab;PDBTitle: crystal structure of the molybdopterin biosynthesis enzyme moab2 (ttha0341) from thermus theromophilus hb8
14 d1y5ea1
85.8
7
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MogA-like15 d1xi8a3
84.9
7
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MoeA central domain-like16 c2nqqA_
84.8
12
PDB header: biosynthetic proteinChain: A: PDB Molecule: molybdopterin biosynthesis protein moea;PDBTitle: moea r137q
17 c2fu3A_
84.6
16
PDB header: biosynthetic protein/structural proteinChain: A: PDB Molecule: gephyrin;PDBTitle: crystal structure of gephyrin e-domain
18 c2pjkA_
83.2
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: 178aa long hypothetical molybdenum cofactorPDBTitle: structure of hypothetical molybdenum cofactor biosynthesis2 protein b from sulfolobus tokodaii
19 d2nqra3
82.1
14
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MoeA central domain-like20 d1jlja_
81.4
19
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MogA-like21 c2e76D_
80.0
11
PDB header: photosynthesisChain: D: PDB Molecule: cytochrome b6-f complex iron-sulfur subunit;PDBTitle: crystal structure of the cytochrome b6f complex with tridecyl-2 stigmatellin (tds) from m.laminosus
22 c2fynO_
78.2
42
PDB header: oxidoreductaseChain: O: PDB Molecule: ubiquinol-cytochrome c reductase iron-sulfurPDBTitle: crystal structure analysis of the double mutant rhodobacter2 sphaeroides bc1 complex
23 c2pq4B_
not modelled
73.8
35
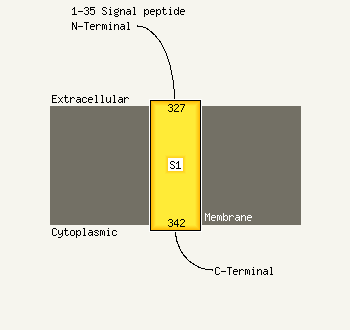
PDB header: chaperone/oxidoreductaseChain: B: PDB Molecule: periplasmic nitrate reductase precursor;PDBTitle: nmr solution structure of napd in complex with napa1-352 signal peptide
24 c1p84E_
73.7
18
PDB header: oxidoreductaseChain: E: PDB Molecule: ubiquinol-cytochrome c reductase iron-sulfurPDBTitle: hdbt inhibited yeast cytochrome bc1 complex
25 d1wu2a3
not modelled
68.5
11
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MoeA central domain-like26 d1uuya_
not modelled
68.4
12
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MogA-like27 d1uz5a3
not modelled
65.1
16
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MoeA central domain-like28 d1mkza_
not modelled
60.8
12
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MogA-like29 c3rfqC_
not modelled
60.1
9
PDB header: biosynthetic proteinChain: C: PDB Molecule: pterin-4-alpha-carbinolamine dehydratase moab2;PDBTitle: crystal structure of pterin-4-alpha-carbinolamine dehydratase moab22 from mycobacterium marinum
30 c2wcvI_
not modelled
59.8
27
PDB header: isomeraseChain: I: PDB Molecule: l-fucose mutarotase;PDBTitle: crystal structure of bacterial fucu
31 c2fyuE_
not modelled
57.2
14
PDB header: oxidoreductaseChain: E: PDB Molecule: ubiquinol-cytochrome c reductase iron-sulfur subunit,PDBTitle: crystal structure of bovine heart mitochondrial bc1 with jg1442 inhibitor
32 d2h1qa1
not modelled
56.7
9
Fold: PLP-dependent transferase-likeSuperfamily: Dhaf3308-likeFamily: Dhaf3308-like33 d1ogda_
not modelled
53.2
23
Fold: RbsD-likeSuperfamily: RbsD-likeFamily: RbsD-like34 c1uz5A_
not modelled
52.7
14
PDB header: molybdopterin biosynthesisChain: A: PDB Molecule: 402aa long hypothetical molybdopterinPDBTitle: the crystal structure of molybdopterin biosynthesis moea2 protein from pyrococcus horikosii
35 c3e7nB_
not modelled
52.3
31
PDB header: transport proteinChain: B: PDB Molecule: d-ribose high-affinity transport system;PDBTitle: crystal structure of d-ribose high-affinity transport system from2 salmonella typhimurium lt2
36 d1ovma1
not modelled
52.1
26
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain37 c3mvkA_
not modelled
45.9
23
PDB header: isomeraseChain: A: PDB Molecule: protein fucu;PDBTitle: the crystal structure of fucu from bifidobacterium longum to 1.65a
38 d2ob5a1
not modelled
44.0
23
Fold: RbsD-likeSuperfamily: RbsD-likeFamily: RbsD-like39 d1pvda1
not modelled
42.3
18
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain40 c2wcuB_
not modelled
41.9
27
PDB header: isomeraseChain: B: PDB Molecule: protein fucu homolog;PDBTitle: crystal structure of mammalian fucu
41 c3ilhA_
not modelled
40.0
14
PDB header: transcription regulatorChain: A: PDB Molecule: two component response regulator;PDBTitle: crystal structure of two component response regulator from cytophaga2 hutchinsonii
42 c3a52A_
not modelled
38.9
21
PDB header: hydrolaseChain: A: PDB Molecule: cold-active alkaline phosphatase;PDBTitle: crystal structure of cold-active alkailne phosphatase from2 psychrophile shewanella sp.
43 d2oa4a1
not modelled
35.6
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: TrpR-likeFamily: SPO1678-like44 d1k7ha_
not modelled
34.0
29
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Alkaline phosphatase45 d1whza_
not modelled
33.7
27
Fold: dsRBD-likeSuperfamily: YcfA/nrd intein domainFamily: YcfA-like46 d1y6va1
not modelled
33.6
25
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Alkaline phosphatase47 d2f7wa1
not modelled
33.4
10
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MogA-like48 d2g2ca1
not modelled
33.4
12
Fold: Molybdenum cofactor biosynthesis proteinsSuperfamily: Molybdenum cofactor biosynthesis proteinsFamily: MogA-like49 d1vmea1
not modelled
32.8
16
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related50 c3e2dB_
not modelled
30.8
21
PDB header: hydrolaseChain: B: PDB Molecule: alkaline phosphatase;PDBTitle: the 1.4 a crystal structure of the large and cold-active2 vibrio sp. alkaline phosphatase
51 d1zpda1
not modelled
30.1
18
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain52 c2x98A_
not modelled
29.7
17
PDB header: hydrolaseChain: A: PDB Molecule: alkaline phosphatase;PDBTitle: h.salinarum alkaline phosphatase
53 c3fniA_
not modelled
29.1
11
PDB header: oxidoreductaseChain: A: PDB Molecule: putative diflavin flavoprotein a 3;PDBTitle: crystal structure of a diflavin flavoprotein a3 (all3895) from nostoc2 sp., northeast structural genomics consortium target nsr431a
54 d1ozha1
not modelled
28.6
18
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain55 c1ew2A_
not modelled
26.3
25
PDB header: hydrolaseChain: A: PDB Molecule: phosphatase;PDBTitle: crystal structure of a human phosphatase
56 d1zeda1
not modelled
26.3
25
Fold: Alkaline phosphatase-likeSuperfamily: Alkaline phosphatase-likeFamily: Alkaline phosphatase57 d1xhja_
not modelled
26.2
14
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: NifU C-terminal domain-like58 c2jnvA_
not modelled
25.4
22
PDB header: metal transportChain: A: PDB Molecule: nifu-like protein 1, chloroplast;PDBTitle: solution structure of c-terminal domain of nifu-like2 protein from oryza sativa
59 c2iucB_
not modelled
24.9
8
PDB header: hydrolaseChain: B: PDB Molecule: alkaline phosphatase;PDBTitle: structure of alkaline phosphatase from the antarctic2 bacterium tab5
60 c2jrtA_
not modelled
24.5
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: nmr solution structure of the protein coded by gene2 rhos4_12090 of rhodobacter sphaeroides. northeast3 structural genomics target rhr5
61 d1ik6a2
not modelled
24.2
12
Fold: TK C-terminal domain-likeSuperfamily: TK C-terminal domain-likeFamily: Branched-chain alpha-keto acid dehydrogenase beta-subunit, C-terminal-domain62 c1wu2B_
not modelled
23.9
19
PDB header: structural genomics,biosynthetic proteinChain: B: PDB Molecule: molybdopterin biosynthesis moea protein;PDBTitle: crystal structure of molybdopterin biosynthesis moea2 protein from pyrococcus horikoshii ot3
63 c3hbcA_
not modelled
22.4
12
PDB header: hydrolaseChain: A: PDB Molecule: choloylglycine hydrolase;PDBTitle: crystal structure of choloylglycine hydrolase from bacteroides2 thetaiotaomicron vpi
64 c3fq6A_
not modelled
22.3
16
PDB header: transferaseChain: A: PDB Molecule: methyltransferase;PDBTitle: the crystal structure of a methyltransferase domain from bacteroides2 thetaiotaomicron vpi
65 d1rlja_
not modelled
21.2
15
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavoprotein NrdI66 d1ryda1
not modelled
19.7
14
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain67 d2djia1
not modelled
19.6
8
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain68 c3uhjE_
not modelled
19.2
15
PDB header: oxidoreductaseChain: E: PDB Molecule: probable glycerol dehydrogenase;PDBTitle: crystal structure of a probable glycerol dehydrogenase from2 sinorhizobium meliloti 1021
69 c2zwmA_
not modelled
18.7
17
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulatory protein yycf;PDBTitle: crystal structure of yycf receiver domain from bacillus2 subtilis
70 d2ez9a1
not modelled
18.6
16
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain71 c2oasA_
not modelled
18.2
23
PDB header: transferaseChain: A: PDB Molecule: 4-hydroxybutyrate coenzyme a transferase;PDBTitle: crystal structure of 4-hydroxybutyrate coenzyme a transferase (atoa)2 in complex with coa from shewanella oneidensis, northeast structural3 genomics target sor119.
72 c3nhzA_
not modelled
18.1
17
PDB header: dna binding proteinChain: A: PDB Molecule: two component system transcriptional regulator mtra;PDBTitle: structure of n-terminal domain of mtra
73 c2kveA_
not modelled
17.1
67
PDB header: hormoneChain: A: PDB Molecule: mesencephalic astrocyte-derived neurotrophic factor;PDBTitle: c-terminal domain of mesencephalic astrocyte-derived neurotrophic2 factor (manf)
74 d1t9ba1
not modelled
17.0
22
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain75 d1zh2a1
not modelled
16.2
12
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related76 c3up8B_
not modelled
15.9
13
PDB header: oxidoreductaseChain: B: PDB Molecule: putative 2,5-diketo-d-gluconic acid reductase b;PDBTitle: crystal structure of a putative 2,5-diketo-d-gluconic acid reductase b
77 d1kgsa2
not modelled
15.8
14
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related78 d2f6ma1
not modelled
15.6
18
Fold: Long alpha-hairpinSuperfamily: Endosomal sorting complex assembly domainFamily: VPS23 C-terminal domain79 c3p13B_
not modelled
15.6
19
PDB header: isomeraseChain: B: PDB Molecule: d-ribose pyranase;PDBTitle: complex structure of d-ribose pyranase sa240 with d-ribose
80 d1ys7a2
not modelled
15.5
12
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related81 d1ofcx1
not modelled
15.4
23
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Myb/SANT domain82 c2qvgA_
not modelled
14.3
21
PDB header: transferaseChain: A: PDB Molecule: two component response regulator;PDBTitle: the crystal structure of a two-component response regulator2 from legionella pneumophila
83 d1uhua_
not modelled
13.7
18
Fold: Retroviral matrix proteinsSuperfamily: Retroviral matrix proteinsFamily: MMLV matrix protein-like84 d1evsa_
not modelled
13.2
33
Fold: 4-helical cytokinesSuperfamily: 4-helical cytokinesFamily: Long-chain cytokines85 c3eh7A_
not modelled
13.1
21
PDB header: transferaseChain: A: PDB Molecule: 4-hydroxybutyrate coa-transferase;PDBTitle: the structure of a putative 4-hydroxybutyrate coa-transferase from2 porphyromonas gingivalis w83
86 d1peya_
not modelled
13.1
23
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related87 d1xhfa1
not modelled
12.9
15
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related88 c1ovmC_
not modelled
12.7
26
PDB header: lyaseChain: C: PDB Molecule: indole-3-pyruvate decarboxylase;PDBTitle: crystal structure of indolepyruvate decarboxylase from2 enterobacter cloacae
89 c2w0yB_
not modelled
12.7
12
PDB header: hydrolaseChain: B: PDB Molecule: alkaline phosphatase;PDBTitle: h.salinarum alkaline phosphatase
90 c3kcnA_
not modelled
12.6
18
PDB header: lyaseChain: A: PDB Molecule: adenylate cyclase homolog;PDBTitle: the crystal structure of adenylate cyclase from2 rhodopirellula baltica
91 c2r8bA_
not modelled
12.3
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein atu2452;PDBTitle: the crystal structure of the protein atu2452 of unknown function from2 agrobacterium tumefaciens str. c58
92 d1hn4a_
not modelled
12.3
16
Fold: Phospholipase A2, PLA2Superfamily: Phospholipase A2, PLA2Family: Vertebrate phospholipase A293 d1ycga1
not modelled
11.7
11
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: Flavodoxin-related94 d1ny5a1
not modelled
11.6
16
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related95 c3d7rB_
not modelled
11.6
8
PDB header: hydrolaseChain: B: PDB Molecule: esterase;PDBTitle: crystal structure of a putative esterase from staphylococcus aureus
96 c3eq2A_
not modelled
11.6
20
PDB header: signaling proteinChain: A: PDB Molecule: probable two-component response regulator;PDBTitle: structure of hexagonal crystal form of pseudomonas2 aeruginosa rssb
97 d1veha_
not modelled
11.5
33
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: NifU C-terminal domain-like98 d2r25b1
not modelled
11.4
13
Fold: Flavodoxin-likeSuperfamily: CheY-likeFamily: CheY-related99 c3luaA_
not modelled
11.2
11
PDB header: transcription regulatorChain: A: PDB Molecule: response regulator receiver protein;PDBTitle: crystal structure of a signal receiver domain of two component signal2 transduction (histidine kinase) from clostridium thermocellum