| 1 |
|
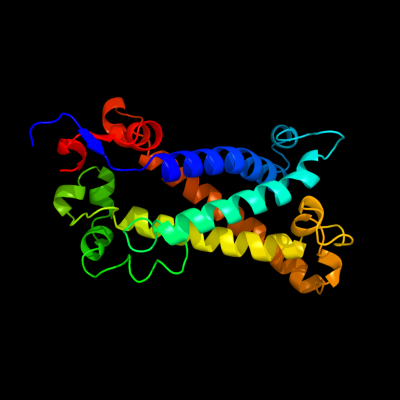
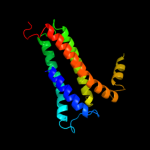
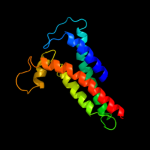
PDB 1kqf chain C
Region: 6 - 228
Aligned: 193
Modelled: 208
Confidence: 100.0%
Identity: 16%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Formate dehydrogenase N, cytochrome (gamma) subunit
Phyre2
| 2 |
|
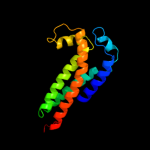
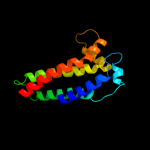
PDB 2qjk chain M
Region: 24 - 213
Aligned: 155
Modelled: 155
Confidence: 98.3%
Identity: 15%
PDB header:electron transport
Chain: M: PDB Molecule:cytochrome b;
PDBTitle: crystal structure analysis of mutant rhodobacter2 sphaeroides bc1 with stigmatellin and antimycin
Phyre2
| 3 |
|
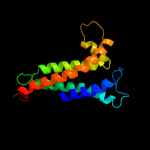
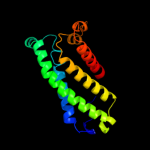
PDB 3cx5 chain C domain 2
Region: 24 - 213
Aligned: 155
Modelled: 155
Confidence: 98.3%
Identity: 17%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 4 |
|
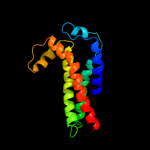
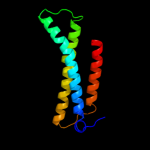
PDB 3cx5 chain N
Region: 24 - 213
Aligned: 155
Modelled: 155
Confidence: 98.2%
Identity: 17%
PDB header:oxidoreductase
Chain: N: PDB Molecule:cytochrome b;
PDBTitle: structure of complex iii with bound cytochrome c in reduced2 state and definition of a minimal core interface for3 electron transfer.
Phyre2
| 5 |
|
PDB 3cwb chain C
Region: 24 - 213
Aligned: 153
Modelled: 154
Confidence: 98.2%
Identity: 16%
PDB header:oxidoreductase
Chain: C: PDB Molecule:cytochrome b;
PDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d
Phyre2
| 6 |
|
PDB 1ppj chain C domain 2
Region: 24 - 213
Aligned: 153
Modelled: 154
Confidence: 98.1%
Identity: 18%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 7 |
|
PDB 1q90 chain B
Region: 24 - 205
Aligned: 147
Modelled: 147
Confidence: 98.0%
Identity: 18%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 8 |
|
PDB 1bcc chain C domain 3
Region: 24 - 205
Aligned: 145
Modelled: 146
Confidence: 98.0%
Identity: 17%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 9 |
|
PDB 2e74 chain A domain 1
Region: 24 - 205
Aligned: 147
Modelled: 147
Confidence: 97.9%
Identity: 18%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 10 |
|
PDB 1y5i chain C domain 1
Region: 1 - 196
Aligned: 147
Modelled: 158
Confidence: 84.7%
Identity: 7%
Fold: Heme-binding four-helical bundle
Superfamily: Respiratory nitrate reductase 1 gamma chain
Family: Respiratory nitrate reductase 1 gamma chain
Phyre2
| 11 |
|
PDB 3eh4 chain A
Region: 6 - 148
Aligned: 111
Modelled: 111
Confidence: 53.0%
Identity: 14%
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome c oxidase subunit 1;
PDBTitle: structure of the reduced form of cytochrome ba3 oxidase from thermus2 thermophilus
Phyre2
| 12 |
|
PDB 2axt chain A domain 1
Region: 123 - 228
Aligned: 105
Modelled: 106
Confidence: 16.0%
Identity: 14%
Fold: Bacterial photosystem II reaction centre, L and M subunits
Superfamily: Bacterial photosystem II reaction centre, L and M subunits
Family: Bacterial photosystem II reaction centre, L and M subunits
Phyre2