| 1 |
|
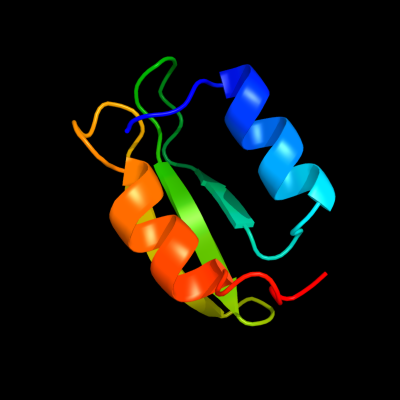
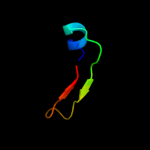
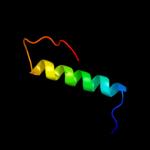
PDB 2hjj chain A domain 1
Region: 16 - 78
Aligned: 63
Modelled: 63
Confidence: 100.0%
Identity: 57%
Fold: dsRBD-like
Superfamily: YcfA/nrd intein domain
Family: YkfF-like
Phyre2
| 2 |
|
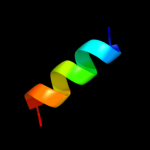
PDB 2hjj chain A
Region: 16 - 78
Aligned: 63
Modelled: 63
Confidence: 100.0%
Identity: 57%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein ykff;
PDBTitle: solution nmr structure of protein ykff from escherichia coli.2 northeast structural genomics target er397.
Phyre2
| 3 |
|
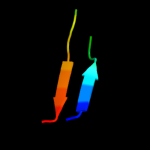
PDB 1xs3 chain A
Region: 33 - 50
Aligned: 18
Modelled: 18
Confidence: 13.4%
Identity: 22%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein xc975;
PDBTitle: solution structure analysis of the xc975 protein
Phyre2
| 4 |
|
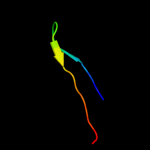
PDB 1iio chain A
Region: 9 - 23
Aligned: 15
Modelled: 15
Confidence: 12.9%
Identity: 33%
Fold: EF Hand-like
Superfamily: Hypothetical protein MTH865
Family: Hypothetical protein MTH865
Phyre2
| 5 |
|
PDB 3isy chain A
Region: 40 - 56
Aligned: 17
Modelled: 17
Confidence: 11.7%
Identity: 35%
PDB header:protein binding
Chain: A: PDB Molecule:intracellular proteinase inhibitor;
PDBTitle: crystal structure of an intracellular proteinase inhibitor (ipi,2 bsu11130) from bacillus subtilis at 2.61 a resolution
Phyre2
| 6 |
|
PDB 3tr3 chain A
Region: 33 - 50
Aligned: 18
Modelled: 18
Confidence: 11.0%
Identity: 28%
PDB header:unknown function
Chain: A: PDB Molecule:bola;
PDBTitle: structure of a bola protein homologue from coxiella burnetii
Phyre2
| 7 |
|
PDB 1yew chain F
Region: 65 - 76
Aligned: 12
Modelled: 12
Confidence: 9.5%
Identity: 33%
PDB header:oxidoreductase, membrane protein
Chain: F: PDB Molecule:particulate methane monooxygenase, a subunit;
PDBTitle: crystal structure of particulate methane monooxygenase
Phyre2
| 8 |
|
PDB 1r1q chain A
Region: 1 - 71
Aligned: 70
Modelled: 71
Confidence: 9.1%
Identity: 13%
Fold: SH2-like
Superfamily: SH2 domain
Family: SH2 domain
Phyre2
| 9 |
|
PDB 2dhm chain A
Region: 21 - 47
Aligned: 27
Modelled: 27
Confidence: 9.0%
Identity: 19%
PDB header:protein binding
Chain: A: PDB Molecule:protein bola;
PDBTitle: solution structure of the bola protein from escherichia coli
Phyre2
| 10 |
|
PDB 2zt9 chain F
Region: 64 - 77
Aligned: 14
Modelled: 14
Confidence: 8.1%
Identity: 29%
PDB header:photosynthesis
Chain: F: PDB Molecule:cytochrome b6-f complex subunit 7;
PDBTitle: crystal structure of the cytochrome b6f complex from nostoc sp. pcc2 7120
Phyre2
| 11 |
|
PDB 3o2e chain A
Region: 33 - 47
Aligned: 15
Modelled: 15
Confidence: 8.1%
Identity: 20%
PDB header:unknown function
Chain: A: PDB Molecule:bola-like protein;
PDBTitle: crystal structure of a bol-like protein from babesia bovis
Phyre2
| 12 |
|
PDB 2kdn chain A
Region: 33 - 47
Aligned: 15
Modelled: 15
Confidence: 7.6%
Identity: 27%
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein pfe0790c;
PDBTitle: solution structure of pfe0790c, a putative bola-like2 protein from the protozoan parasite plasmodium falciparum.
Phyre2
| 13 |
|
PDB 2q2e chain B
Region: 32 - 51
Aligned: 20
Modelled: 20
Confidence: 7.6%
Identity: 20%
PDB header:isomerase
Chain: B: PDB Molecule:type 2 dna topoisomerase 6 subunit b;
PDBTitle: crystal structure of the topoisomerase vi holoenzyme from2 methanosarcina mazei
Phyre2
| 14 |
|
PDB 1rwh chain A domain 3
Region: 44 - 78
Aligned: 35
Modelled: 35
Confidence: 7.2%
Identity: 23%
Fold: Supersandwich
Superfamily: Galactose mutarotase-like
Family: Hyaluronate lyase-like, central domain
Phyre2
| 15 |
|
PDB 3chx chain F
Region: 65 - 76
Aligned: 12
Modelled: 12
Confidence: 7.0%
Identity: 33%
PDB header:membrane protein
Chain: F: PDB Molecule:pmoa;
PDBTitle: crystal structure of methylosinus trichosporium ob3b2 particulate methane monooxygenase (pmmo)
Phyre2
| 16 |
|
PDB 1k1s chain A domain 1
Region: 16 - 46
Aligned: 31
Modelled: 31
Confidence: 6.6%
Identity: 16%
Fold: Lesion bypass DNA polymerase (Y-family), little finger domain
Superfamily: Lesion bypass DNA polymerase (Y-family), little finger domain
Family: Lesion bypass DNA polymerase (Y-family), little finger domain
Phyre2
| 17 |
|
PDB 1v9j chain A
Region: 33 - 47
Aligned: 15
Modelled: 15
Confidence: 5.3%
Identity: 40%
Fold: Alpha-lytic protease prodomain-like
Superfamily: BolA-like
Family: BolA-like
Phyre2
| 18 |
|
PDB 1f1s chain A domain 4
Region: 44 - 78
Aligned: 35
Modelled: 35
Confidence: 5.2%
Identity: 20%
Fold: Supersandwich
Superfamily: Galactose mutarotase-like
Family: Hyaluronate lyase-like, central domain
Phyre2
| 19 |
|
PDB 2zgw chain A domain 1
Region: 44 - 54
Aligned: 11
Modelled: 11
Confidence: 5.2%
Identity: 36%
Fold: SH3-like barrel
Superfamily: C-terminal domain of transcriptional repressors
Family: Biotin repressor (BirA)
Phyre2