| 1 |
|
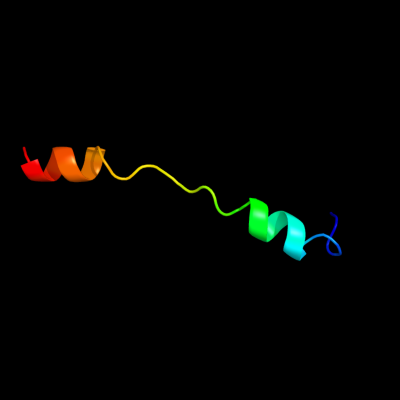
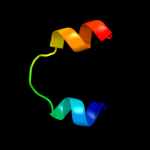
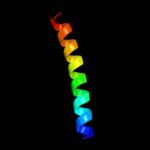
PDB 2x2v chain G
Region: 10 - 37
Aligned: 28
Modelled: 28
Confidence: 40.4%
Identity: 39%
PDB header:membrane protein
Chain: G: PDB Molecule:atp synthase subunit c;
PDBTitle: structural basis of a novel proton-coordination type in an2 f1fo-atp synthase rotor ring
Phyre2
| 2 |
|
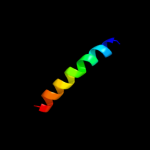
PDB 1yce chain D
Region: 10 - 37
Aligned: 28
Modelled: 28
Confidence: 32.0%
Identity: 36%
PDB header:membrane protein
Chain: D: PDB Molecule:subunit c;
PDBTitle: structure of the rotor ring of f-type na+-atpase from ilyobacter2 tartaricus
Phyre2
| 3 |
|
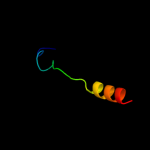
PDB 2w5j chain M
Region: 10 - 37
Aligned: 28
Modelled: 28
Confidence: 30.9%
Identity: 39%
PDB header:hydrolase
Chain: M: PDB Molecule:atp synthase c chain, chloroplastic;
PDBTitle: structure of the c14-rotor ring of the proton translocating2 chloroplast atp synthase
Phyre2
| 4 |
|
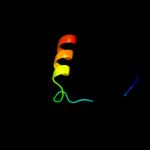
PDB 2wpd chain P
Region: 10 - 37
Aligned: 28
Modelled: 28
Confidence: 29.3%
Identity: 39%
PDB header:hydrolase
Chain: P: PDB Molecule:atp synthase subunit 9, mitochondrial;
PDBTitle: the mg.adp inhibited state of the yeast f1c10 atp synthase
Phyre2
| 5 |
|
PDB 1shz chain F
Region: 16 - 35
Aligned: 20
Modelled: 20
Confidence: 23.6%
Identity: 35%
PDB header:signaling protein
Chain: F: PDB Molecule:rho guanine nucleotide exchange factor 1;
PDBTitle: crystal structure of the p115rhogef rgrgs domain in a2 complex with galpha(13):galpha(i1) chimera
Phyre2
| 6 |
|
PDB 1wu0 chain A
Region: 10 - 28
Aligned: 19
Modelled: 19
Confidence: 21.6%
Identity: 42%
PDB header:hydrolase
Chain: A: PDB Molecule:atp synthase c chain;
PDBTitle: solution structure of subunit c of f1fo-atp synthase from2 the thermophilic bacillus ps3
Phyre2
| 7 |
|
PDB 1htj chain F
Region: 15 - 35
Aligned: 21
Modelled: 21
Confidence: 20.5%
Identity: 38%
Fold: Regulator of G-protein signaling, RGS
Superfamily: Regulator of G-protein signaling, RGS
Family: Regulator of G-protein signaling, RGS
Phyre2
| 8 |
|
PDB 1htj chain F
Region: 15 - 35
Aligned: 21
Modelled: 21
Confidence: 20.5%
Identity: 38%
PDB header:signaling protein
Chain: F: PDB Molecule:kiaa0380;
PDBTitle: structure of the rgs-like domain from pdz-rhogef
Phyre2
| 9 |
|
PDB 1iap chain A
Region: 16 - 35
Aligned: 20
Modelled: 20
Confidence: 18.9%
Identity: 35%
Fold: Regulator of G-protein signaling, RGS
Superfamily: Regulator of G-protein signaling, RGS
Family: Regulator of G-protein signaling, RGS
Phyre2
| 10 |
|
PDB 1ceu chain A
Region: 2 - 12
Aligned: 11
Modelled: 11
Confidence: 17.4%
Identity: 36%
PDB header:viral protein
Chain: A: PDB Molecule:protein (hiv-1 regulatory protein n-terminal
PDBTitle: nmr structure of the (1-51) n-terminal domain of the hiv-12 regulatory protein
Phyre2
| 11 |
|
PDB 2lat chain A
Region: 16 - 40
Aligned: 25
Modelled: 25
Confidence: 10.2%
Identity: 36%
PDB header:membrane protein
Chain: A: PDB Molecule:dolichyl-diphosphooligosaccharide--protein
PDBTitle: solution structure of a human minimembrane protein ost4
Phyre2
| 12 |
|
PDB 1xme chain B domain 2
Region: 16 - 38
Aligned: 21
Modelled: 23
Confidence: 9.6%
Identity: 43%
Fold: Transmembrane helix hairpin
Superfamily: Cytochrome c oxidase subunit II-like, transmembrane region
Family: Cytochrome c oxidase subunit II-like, transmembrane region
Phyre2
| 13 |
|
PDB 2xnd chain K
Region: 10 - 37
Aligned: 28
Modelled: 28
Confidence: 8.1%
Identity: 43%
PDB header:hydrolase
Chain: K: PDB Molecule:atp synthase lipid-binding protein, mitochondrial;
PDBTitle: crystal structure of bovine f1-c8 sub-complex of atp2 synthase
Phyre2
| 14 |
|
PDB 3bc1 chain F
Region: 9 - 17
Aligned: 9
Modelled: 9
Confidence: 8.1%
Identity: 56%
PDB header:signaling protein/transport protein
Chain: F: PDB Molecule:synaptotagmin-like protein 2;
PDBTitle: crystal structure of the complex rab27a-slp2a
Phyre2
| 15 |
|
PDB 1m8l chain A
Region: 2 - 18
Aligned: 17
Modelled: 17
Confidence: 8.0%
Identity: 47%
PDB header:viral protein
Chain: A: PDB Molecule:vpr protein;
PDBTitle: nmr structure of the hiv-1 regulatory protein vpr
Phyre2
| 16 |
|
PDB 1sg7 chain A
Region: 4 - 22
Aligned: 19
Modelled: 19
Confidence: 8.0%
Identity: 42%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative cation transport regulator chab;
PDBTitle: nmr solution structure of the putative cation transport2 regulator chab
Phyre2
| 17 |
|
PDB 1sg7 chain A domain 1
Region: 4 - 22
Aligned: 19
Modelled: 19
Confidence: 8.0%
Identity: 42%
Fold: ChaB-like
Superfamily: ChaB-like
Family: ChaB-like
Phyre2
| 18 |
|
PDB 3h87 chain D
Region: 11 - 19
Aligned: 9
Modelled: 9
Confidence: 7.6%
Identity: 78%
PDB header:toxin/antitoxin
Chain: D: PDB Molecule:putative uncharacterized protein;
PDBTitle: rv0301 rv0300 toxin antitoxin complex from mycobacterium tuberculosis
Phyre2
| 19 |
|
PDB 3mwz chain A
Region: 1 - 22
Aligned: 22
Modelled: 22
Confidence: 7.5%
Identity: 27%
PDB header:hydrolase inhibitor
Chain: A: PDB Molecule:sialostatin l2;
PDBTitle: crystal structure of the selenomethionine derivative of the l 22,47,2 100 m mutant of sialostatin l2
Phyre2
| 20 |
|
PDB 1nek chain C
Region: 12 - 38
Aligned: 27
Modelled: 27
Confidence: 6.8%
Identity: 15%
Fold: Heme-binding four-helical bundle
Superfamily: Fumarate reductase respiratory complex transmembrane subunits
Family: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)
Phyre2
| 21 |
|
| 22 |
|