| 1 |
|
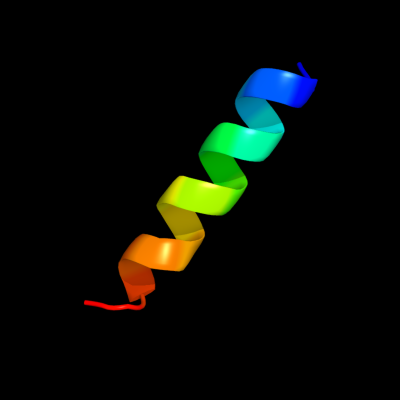
PDB 2hg5 chain D
Region: 18 - 35
Aligned: 18
Modelled: 18
Confidence: 10.4%
Identity: 17%
PDB header:membrane protein
Chain: D: PDB Molecule:kcsa channel;
PDBTitle: cs+ complex of a k channel with an amide to ester substitution in the2 selectivity filter
Phyre2
| 2 |
|
PDB 3t41 chain B
Region: 14 - 27
Aligned: 14
Modelled: 14
Confidence: 6.4%
Identity: 36%
PDB header:hydrolase
Chain: B: PDB Molecule:epidermin leader peptide processing serine protease epip;
PDBTitle: 1.95 angstrom resolution crystal structure of epidermin leader peptide2 processing serine protease (epip) s393a mutant from staphylococcus3 aureus
Phyre2
| 3 |
|
PDB 1r0r chain E
Region: 14 - 27
Aligned: 14
Modelled: 14
Confidence: 6.3%
Identity: 29%
Fold: Subtilisin-like
Superfamily: Subtilisin-like
Family: Subtilases
Phyre2
| 4 |
|
PDB 3lpc chain A
Region: 14 - 27
Aligned: 14
Modelled: 14
Confidence: 5.7%
Identity: 29%
PDB header:hydrolase
Chain: A: PDB Molecule:aprb2;
PDBTitle: crystal structure of a subtilisin-like protease
Phyre2
| 5 |
|
PDB 1s2n chain B
Region: 14 - 27
Aligned: 14
Modelled: 14
Confidence: 5.6%
Identity: 29%
PDB header:hydrolase
Chain: B: PDB Molecule:extracellular subtilisin-like serine proteinase;
PDBTitle: crystal strucure of a cold adapted subtilisin-like serine proteinase
Phyre2
| 6 |
|
PDB 2b6n chain A
Region: 14 - 27
Aligned: 14
Modelled: 14
Confidence: 5.6%
Identity: 29%
PDB header:hydrolase
Chain: A: PDB Molecule:proteinase k;
PDBTitle: the 1.8 a crystal structure of a proteinase k like enzyme from a2 psychrotroph serratia species
Phyre2
| 7 |
|
PDB 1loi chain A
Region: 27 - 35
Aligned: 9
Modelled: 9
Confidence: 5.5%
Identity: 56%
PDB header:hydrolase
Chain: A: PDB Molecule:cyclic 3',5'-amp specific phosphodiesterase rd1;
PDBTitle: n-terminal splice region of rat c-amp phosphodiesterase,2 nmr, 7 structures
Phyre2
| 8 |
|
PDB 2iy9 chain A
Region: 15 - 27
Aligned: 13
Modelled: 13
Confidence: 5.3%
Identity: 31%
PDB header:toxin
Chain: A: PDB Molecule:suba;
PDBTitle: crystal structure of the a-subunit of the ab5 toxin from e.2 coli
Phyre2
| 9 |
|
PDB 1xx7 chain A
Region: 6 - 15
Aligned: 10
Modelled: 10
Confidence: 5.3%
Identity: 30%
Fold: HD-domain/PDEase-like
Superfamily: HD-domain/PDEase-like
Family: HD domain
Phyre2