| 1 |
|
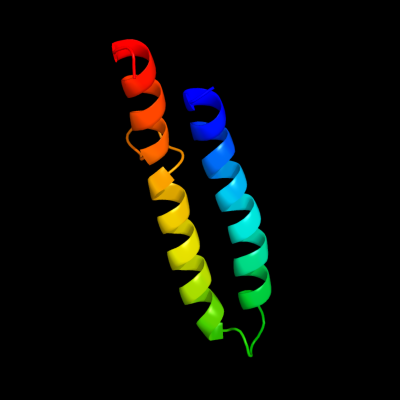
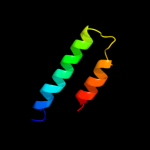
PDB 1wu0 chain A
Region: 84 - 147
Aligned: 61
Modelled: 61
Confidence: 57.6%
Identity: 25%
PDB header:hydrolase
Chain: A: PDB Molecule:atp synthase c chain;
PDBTitle: solution structure of subunit c of f1fo-atp synthase from2 the thermophilic bacillus ps3
Phyre2
| 2 |
|
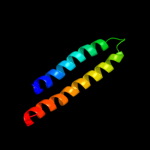
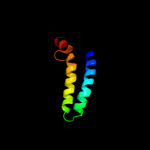
PDB 2x2v chain G
Region: 84 - 126
Aligned: 43
Modelled: 43
Confidence: 14.9%
Identity: 26%
PDB header:membrane protein
Chain: G: PDB Molecule:atp synthase subunit c;
PDBTitle: structural basis of a novel proton-coordination type in an2 f1fo-atp synthase rotor ring
Phyre2
| 3 |
|
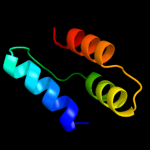
PDB 3dd7 chain A
Region: 35 - 85
Aligned: 51
Modelled: 51
Confidence: 13.0%
Identity: 12%
PDB header:ribosome inhibitor
Chain: A: PDB Molecule:death on curing protein;
PDBTitle: structure of doch66y in complex with the c-terminal domain of phd
Phyre2
| 4 |
|
PDB 2l2t chain A
Region: 134 - 141
Aligned: 8
Modelled: 8
Confidence: 10.9%
Identity: 75%
PDB header:membrane protein
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-4;
PDBTitle: solution nmr structure of the erbb4 dimeric membrane domain
Phyre2
| 5 |
|
PDB 1kve chain A
Region: 84 - 102
Aligned: 19
Modelled: 19
Confidence: 9.3%
Identity: 37%
PDB header:toxin
Chain: A: PDB Molecule:smk toxin;
PDBTitle: killer toxin from halotolerant yeast
Phyre2
| 6 |
|
PDB 2w5j chain M
Region: 85 - 126
Aligned: 42
Modelled: 42
Confidence: 7.9%
Identity: 24%
PDB header:hydrolase
Chain: M: PDB Molecule:atp synthase c chain, chloroplastic;
PDBTitle: structure of the c14-rotor ring of the proton translocating2 chloroplast atp synthase
Phyre2
| 7 |
|
PDB 1c99 chain A
Region: 84 - 145
Aligned: 59
Modelled: 62
Confidence: 7.0%
Identity: 29%
Fold: Transmembrane helix hairpin
Superfamily: F1F0 ATP synthase subunit C
Family: F1F0 ATP synthase subunit C
Phyre2
| 8 |
|
PDB 2kjf chain A
Region: 115 - 133
Aligned: 17
Modelled: 19
Confidence: 6.7%
Identity: 47%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:carnocyclin-a;
PDBTitle: the solution structure of the circular bacteriocin2 carnocyclin a (ccla)
Phyre2