| 1 |
|
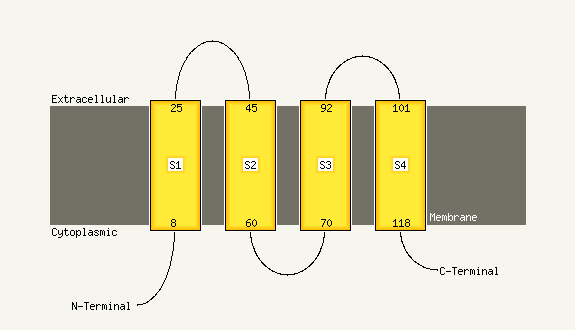
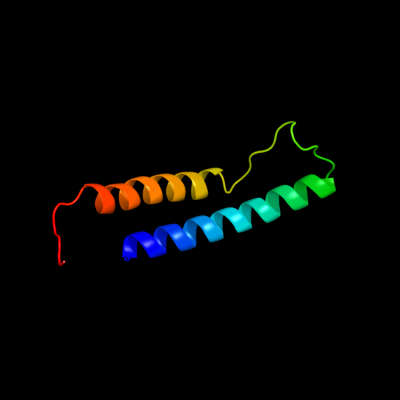
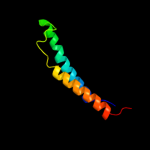
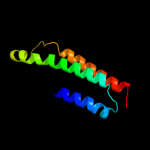
PDB 1xme chain A domain 1
Region: 66 - 131
Aligned: 66
Modelled: 66
Confidence: 57.3%
Identity: 18%
Fold: Cytochrome c oxidase subunit I-like
Superfamily: Cytochrome c oxidase subunit I-like
Family: Cytochrome c oxidase subunit I-like
Phyre2
| 2 |
|
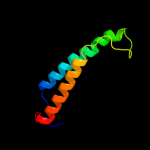
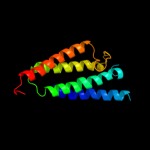
PDB 3dtu chain A domain 1
Region: 59 - 129
Aligned: 71
Modelled: 71
Confidence: 55.7%
Identity: 15%
Fold: Cytochrome c oxidase subunit I-like
Superfamily: Cytochrome c oxidase subunit I-like
Family: Cytochrome c oxidase subunit I-like
Phyre2
| 3 |
|
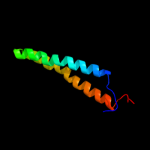
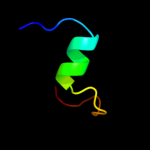
PDB 3eh4 chain A
Region: 66 - 131
Aligned: 66
Modelled: 66
Confidence: 52.4%
Identity: 18%
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome c oxidase subunit 1;
PDBTitle: structure of the reduced form of cytochrome ba3 oxidase from thermus2 thermophilus
Phyre2
| 4 |
|
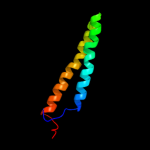
PDB 1ar1 chain A
Region: 62 - 131
Aligned: 70
Modelled: 70
Confidence: 39.2%
Identity: 10%
Fold: Cytochrome c oxidase subunit I-like
Superfamily: Cytochrome c oxidase subunit I-like
Family: Cytochrome c oxidase subunit I-like
Phyre2
| 5 |
|
PDB 1m56 chain G
Region: 59 - 129
Aligned: 71
Modelled: 71
Confidence: 34.4%
Identity: 11%
PDB header:oxidoreductase
Chain: G: PDB Molecule:cytochrome c oxidase;
PDBTitle: structure of cytochrome c oxidase from rhodobactor2 sphaeroides (wild type)
Phyre2
| 6 |
|
PDB 1fft chain A
Region: 59 - 131
Aligned: 73
Modelled: 73
Confidence: 33.0%
Identity: 15%
Fold: Cytochrome c oxidase subunit I-like
Superfamily: Cytochrome c oxidase subunit I-like
Family: Cytochrome c oxidase subunit I-like
Phyre2
| 7 |
|
PDB 1fft chain F
Region: 59 - 131
Aligned: 73
Modelled: 73
Confidence: 33.0%
Identity: 15%
PDB header:oxidoreductase
Chain: F: PDB Molecule:ubiquinol oxidase;
PDBTitle: the structure of ubiquinol oxidase from escherichia coli
Phyre2
| 8 |
|
PDB 1v54 chain A
Region: 43 - 129
Aligned: 87
Modelled: 87
Confidence: 18.6%
Identity: 13%
Fold: Cytochrome c oxidase subunit I-like
Superfamily: Cytochrome c oxidase subunit I-like
Family: Cytochrome c oxidase subunit I-like
Phyre2
| 9 |
|
PDB 3mk7 chain K
Region: 4 - 131
Aligned: 127
Modelled: 128
Confidence: 15.8%
Identity: 17%
PDB header:oxidoreductase
Chain: K: PDB Molecule:cytochrome c oxidase, cbb3-type, subunit n;
PDBTitle: the structure of cbb3 cytochrome oxidase
Phyre2
| 10 |
|
PDB 3pt1 chain A
Region: 81 - 104
Aligned: 24
Modelled: 24
Confidence: 10.7%
Identity: 33%
PDB header:hydrolase
Chain: A: PDB Molecule:upf0364 protein ymr027w;
PDBTitle: structure of duf89 from saccharomyces cerevisiae co-crystallized with2 f6p.
Phyre2