| 1 |
|
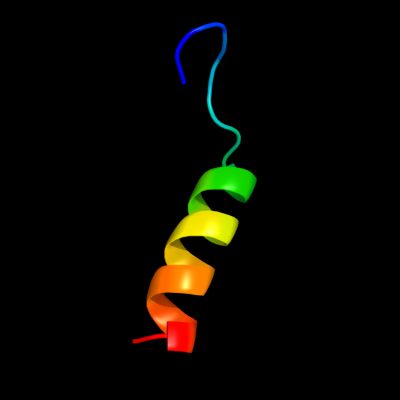
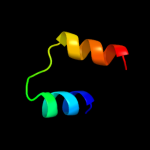
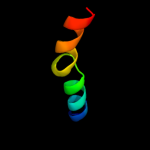
PDB 1jjc chain B domain 4
Region: 29 - 50
Aligned: 22
Modelled: 22
Confidence: 15.3%
Identity: 18%
Fold: Ferredoxin-like
Superfamily: Anticodon-binding domain of PheRS
Family: Anticodon-binding domain of PheRS
Phyre2
| 2 |
|
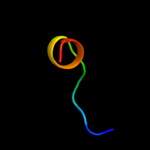
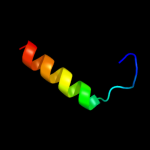
PDB 2owl chain A
Region: 16 - 51
Aligned: 36
Modelled: 36
Confidence: 13.2%
Identity: 19%
PDB header:recombination
Chain: A: PDB Molecule:recombination-associated protein rdgc;
PDBTitle: crystal structure of e. coli rdgc
Phyre2
| 3 |
|
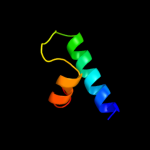
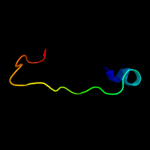
PDB 1usd chain A
Region: 38 - 53
Aligned: 16
Modelled: 16
Confidence: 12.3%
Identity: 25%
PDB header:signaling protein
Chain: A: PDB Molecule:vasodilator-stimulated phosphoprotein;
PDBTitle: human vasp tetramerisation domain l352m
Phyre2
| 4 |
|
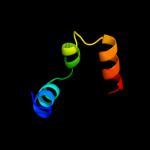
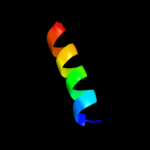
PDB 1ygm chain A
Region: 22 - 47
Aligned: 26
Modelled: 26
Confidence: 11.7%
Identity: 31%
PDB header:membrane protein
Chain: A: PDB Molecule:hypothetical protein bsu31320;
PDBTitle: nmr structure of mistic
Phyre2
| 5 |
|
PDB 1b7y chain B domain 4
Region: 29 - 51
Aligned: 23
Modelled: 23
Confidence: 10.6%
Identity: 17%
Fold: Ferredoxin-like
Superfamily: Anticodon-binding domain of PheRS
Family: Anticodon-binding domain of PheRS
Phyre2
| 6 |
|
PDB 2zc2 chain A
Region: 21 - 46
Aligned: 26
Modelled: 26
Confidence: 9.5%
Identity: 23%
PDB header:replication
Chain: A: PDB Molecule:dnad-like replication protein;
PDBTitle: crystal structure of dnad-like replication protein from2 streptococcus mutans ua159, gi 24377835, residues 127-199
Phyre2
| 7 |
|
PDB 1pu1 chain A
Region: 1 - 13
Aligned: 13
Modelled: 13
Confidence: 9.0%
Identity: 31%
Fold: Hypothetical protein MTH677
Superfamily: Hypothetical protein MTH677
Family: Hypothetical protein MTH677
Phyre2
| 8 |
|
PDB 2r18 chain A
Region: 53 - 88
Aligned: 35
Modelled: 36
Confidence: 6.9%
Identity: 29%
PDB header:viral protein
Chain: A: PDB Molecule:capsid assembly protein vp3;
PDBTitle: structural insights into the multifunctional protein vp3 of2 birnaviruses
Phyre2
| 9 |
|
PDB 2of5 chain K
Region: 11 - 46
Aligned: 36
Modelled: 36
Confidence: 6.2%
Identity: 17%
PDB header:apoptosis
Chain: K: PDB Molecule:leucine-rich repeat and death domain-containing
PDBTitle: oligomeric death domain complex
Phyre2
| 10 |
|
PDB 3bdn chain B
Region: 6 - 31
Aligned: 24
Modelled: 26
Confidence: 6.1%
Identity: 25%
PDB header:transcription/dna
Chain: B: PDB Molecule:lambda repressor;
PDBTitle: crystal structure of the lambda repressor
Phyre2
| 11 |
|
PDB 3cmq chain A
Region: 29 - 51
Aligned: 23
Modelled: 23
Confidence: 5.5%
Identity: 17%
PDB header:ligase
Chain: A: PDB Molecule:phenylalanyl-trna synthetase, mitochondrial;
PDBTitle: crystal structure of human mitochondrial phenylalanine trna2 synthetase
Phyre2
| 12 |
|
PDB 2cb2 chain A domain 1
Region: 24 - 50
Aligned: 27
Modelled: 27
Confidence: 5.5%
Identity: 30%
Fold: Ferredoxin-like
Superfamily: Dimeric alpha+beta barrel
Family: SOR-like
Phyre2
| 13 |
|
PDB 2gid chain P
Region: 74 - 82
Aligned: 9
Modelled: 9
Confidence: 5.3%
Identity: 33%
PDB header:translation
Chain: P: PDB Molecule:mitochondrial rna-binding protein 2;
PDBTitle: crystal structures of trypanosoma bruciei mrp1/mrp2
Phyre2
| 14 |
|
PDB 2owy chain B
Region: 16 - 51
Aligned: 36
Modelled: 36
Confidence: 5.2%
Identity: 19%
PDB header:dna binding protein
Chain: B: PDB Molecule:recombination-associated protein rdgc;
PDBTitle: the recombination-associated protein rdgc adopts a novel toroidal2 architecture for dna binding
Phyre2
| 15 |
|
PDB 1a7g chain E
Region: 62 - 89
Aligned: 27
Modelled: 28
Confidence: 5.1%
Identity: 22%
Fold: Ferredoxin-like
Superfamily: Viral DNA-binding domain
Family: Viral DNA-binding domain
Phyre2
| 16 |
|
PDB 3t97 chain A
Region: 10 - 25
Aligned: 16
Modelled: 16
Confidence: 5.1%
Identity: 31%
PDB header:protein transport
Chain: A: PDB Molecule:nuclear pore glycoprotein p62;
PDBTitle: molecular architecture of the transport channel of the nuclear pore2 complex: nup62/nup54
Phyre2