| 1 |
|
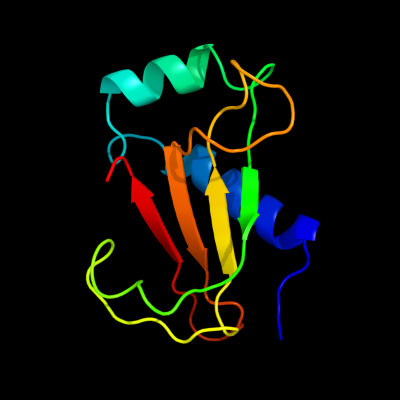
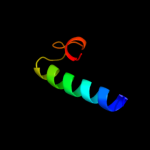
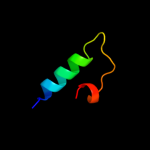
PDB 1nnv chain A
Region: 7 - 109
Aligned: 102
Modelled: 103
Confidence: 100.0%
Identity: 48%
Fold: Cystatin-like
Superfamily: Putative dsDNA mimic
Family: Putative dsDNA mimic
Phyre2
| 2 |
|
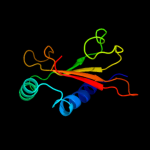
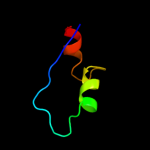
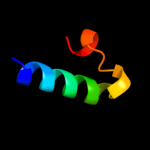
PDB 1bjz chain A
Region: 8 - 36
Aligned: 29
Modelled: 29
Confidence: 14.1%
Identity: 21%
PDB header:transcription regulation
Chain: A: PDB Molecule:tetracycline repressor;
PDBTitle: tetracycline chelated mg2+-ion initiates helix unwinding for tet2 repressor induction
Phyre2
| 3 |
|
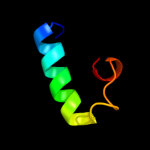
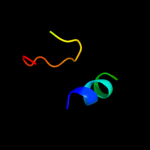
PDB 1bml chain D
Region: 5 - 24
Aligned: 20
Modelled: 20
Confidence: 13.4%
Identity: 30%
PDB header:blood clotting
Chain: D: PDB Molecule:streptokinase;
PDBTitle: complex of the catalytic domain of human plasmin and streptokinase
Phyre2
| 4 |
|
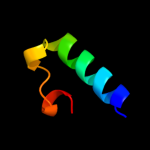
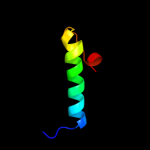
PDB 2np5 chain A domain 1
Region: 10 - 36
Aligned: 27
Modelled: 27
Confidence: 13.4%
Identity: 11%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: Tetracyclin repressor-like, N-terminal domain
Phyre2
| 5 |
|
PDB 1c4p chain C
Region: 6 - 28
Aligned: 23
Modelled: 23
Confidence: 12.1%
Identity: 26%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: Staphylokinase/streptokinase
Family: Staphylokinase/streptokinase
Phyre2
| 6 |
|
PDB 1qqr chain A
Region: 6 - 28
Aligned: 23
Modelled: 23
Confidence: 11.2%
Identity: 26%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: Staphylokinase/streptokinase
Family: Staphylokinase/streptokinase
Phyre2
| 7 |
|
PDB 2g7s chain A domain 1
Region: 6 - 36
Aligned: 31
Modelled: 31
Confidence: 10.8%
Identity: 19%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: Tetracyclin repressor-like, N-terminal domain
Phyre2
| 8 |
|
PDB 2vke chain A domain 1
Region: 8 - 36
Aligned: 29
Modelled: 29
Confidence: 9.3%
Identity: 21%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: Tetracyclin repressor-like, N-terminal domain
Phyre2
| 9 |
|
PDB 1x99 chain A
Region: 69 - 109
Aligned: 35
Modelled: 35
Confidence: 9.0%
Identity: 17%
Fold: Cytolysin/lectin
Superfamily: Cytolysin/lectin
Family: Fungal fruit body lectin
Phyre2
| 10 |
|
PDB 3htx chain A
Region: 82 - 102
Aligned: 21
Modelled: 21
Confidence: 7.2%
Identity: 38%
PDB header:transferase/rna
Chain: A: PDB Molecule:hen1;
PDBTitle: crystal structure of small rna methyltransferase hen1
Phyre2
| 11 |
|
PDB 3o60 chain A
Region: 1 - 36
Aligned: 36
Modelled: 36
Confidence: 7.1%
Identity: 14%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lin0861 protein;
PDBTitle: the crystal structure of lin0861 from listeria innocua to 2.8a
Phyre2
| 12 |
|
PDB 1zk8 chain A domain 1
Region: 9 - 36
Aligned: 28
Modelled: 28
Confidence: 6.8%
Identity: 21%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: Tetracyclin repressor-like, N-terminal domain
Phyre2
| 13 |
|
PDB 2hxi chain A
Region: 8 - 36
Aligned: 29
Modelled: 29
Confidence: 5.7%
Identity: 17%
PDB header:transcription
Chain: A: PDB Molecule:putative transcriptional regulator;
PDBTitle: structural genomics, the crystal structure of a putative2 transcriptional regulator from streptomyces coelicolor a3(2)
Phyre2
| 14 |
|
PDB 2np5 chain A
Region: 10 - 36
Aligned: 27
Modelled: 27
Confidence: 5.3%
Identity: 11%
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator;
PDBTitle: crystal structure of a transcriptional regulator (rha1_ro04179) from2 rhodococcus sp. rha1.
Phyre2