| 1 |
|
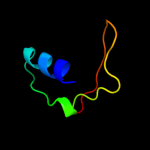
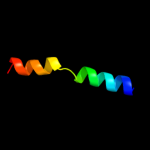
PDB 1avo chain A
Region: 4 - 26
Aligned: 23
Modelled: 23
Confidence: 77.8%
Identity: 30%
PDB header:proteasome activator
Chain: A: PDB Molecule:11s regulator;
PDBTitle: proteasome activator reg(alpha)
Phyre2
| 2 |
|
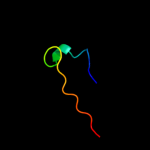
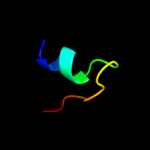
PDB 2a7u chain A
Region: 12 - 26
Aligned: 15
Modelled: 15
Confidence: 42.5%
Identity: 53%
PDB header:hydrolase
Chain: A: PDB Molecule:atp synthase alpha chain;
PDBTitle: nmr solution structure of the e.coli f-atpase delta subunit n-terminal2 domain in complex with alpha subunit n-terminal 22 residues
Phyre2
| 3 |
|
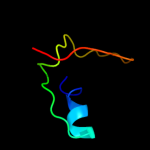
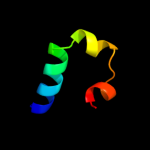
PDB 1div chain A domain 1
Region: 19 - 56
Aligned: 38
Modelled: 38
Confidence: 27.3%
Identity: 21%
Fold: Ribosomal protein L9 C-domain
Superfamily: Ribosomal protein L9 C-domain
Family: Ribosomal protein L9 C-domain
Phyre2
| 4 |
|
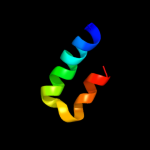
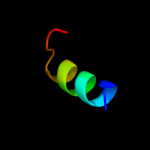
PDB 1uww chain A
Region: 35 - 56
Aligned: 22
Modelled: 22
Confidence: 14.9%
Identity: 41%
Fold: Galactose-binding domain-like
Superfamily: Galactose-binding domain-like
Family: Family 28 carbohydrate binding module, CBM28
Phyre2
| 5 |
|
PDB 2j01 chain I domain 1
Region: 19 - 56
Aligned: 38
Modelled: 38
Confidence: 14.3%
Identity: 24%
Fold: Ribosomal protein L9 C-domain
Superfamily: Ribosomal protein L9 C-domain
Family: Ribosomal protein L9 C-domain
Phyre2
| 6 |
|
PDB 2x43 chain S
Region: 8 - 37
Aligned: 30
Modelled: 30
Confidence: 10.5%
Identity: 37%
PDB header:membrane protein
Chain: S: PDB Molecule:sherp;
PDBTitle: structural basis of molecular recognition by sherp at membrane2 surfaces
Phyre2
| 7 |
|
PDB 2kwy chain A
Region: 3 - 28
Aligned: 26
Modelled: 26
Confidence: 10.1%
Identity: 42%
PDB header:proton transport
Chain: A: PDB Molecule:v-type proton atpase subunit g;
PDBTitle: structure of g61-101
Phyre2
| 8 |
|
PDB 1x6m chain A
Region: 35 - 51
Aligned: 15
Modelled: 17
Confidence: 8.3%
Identity: 60%
Fold: Mss4-like
Superfamily: Mss4-like
Family: Glutathione-dependent formaldehyde-activating enzyme, Gfa
Phyre2
| 9 |
|
PDB 3iwf chain A
Region: 8 - 35
Aligned: 28
Modelled: 28
Confidence: 8.0%
Identity: 25%
PDB header:transcription regulator
Chain: A: PDB Molecule:transcription regulator rpir family;
PDBTitle: the crystal structure of the n-terminal domain of a rpir2 transcriptional regulator from staphylococcus epidermidis to 1.4a
Phyre2
| 10 |
|
PDB 1ybz chain A domain 1
Region: 18 - 33
Aligned: 16
Modelled: 16
Confidence: 6.5%
Identity: 31%
Fold: Chorismate mutase II
Superfamily: Chorismate mutase II
Family: Dimeric chorismate mutase
Phyre2
| 11 |
|
PDB 1rep chain C domain 2
Region: 28 - 43
Aligned: 16
Modelled: 16
Confidence: 6.4%
Identity: 25%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: Replication initiation protein
Phyre2
| 12 |
|
PDB 2nra chain C domain 2
Region: 27 - 39
Aligned: 13
Modelled: 13
Confidence: 6.4%
Identity: 23%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: Replication initiation protein
Phyre2
| 13 |
|
PDB 2qbv chain A
Region: 18 - 33
Aligned: 16
Modelled: 16
Confidence: 5.8%
Identity: 13%
PDB header:isomerase
Chain: A: PDB Molecule:chorismate mutase;
PDBTitle: crystal structure of intracellular chorismate mutase from2 mycobacterium tuberculosis
Phyre2
| 14 |
|
PDB 2k88 chain A
Region: 7 - 27
Aligned: 21
Modelled: 21
Confidence: 5.5%
Identity: 38%
PDB header:hydrolase
Chain: A: PDB Molecule:vacuolar proton pump subunit g;
PDBTitle: association of subunit d (vma6p) and e (vma4p) with g2 (vma10p) and the nmr solution structure of subunit g (g1-3 59) of the saccharomyces cerevisiae v1vo atpase
Phyre2