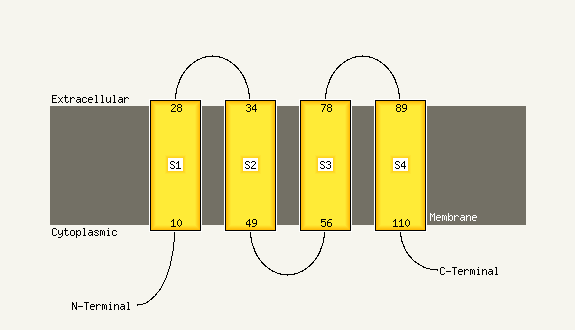
1 c3mk7K_
96.7
14
PDB header: oxidoreductaseChain: K: PDB Molecule: cytochrome c oxidase, cbb3-type, subunit n;PDBTitle: the structure of cbb3 cytochrome oxidase
2 d1ffta_
91.8
18
Fold: Cytochrome c oxidase subunit I-likeSuperfamily: Cytochrome c oxidase subunit I-likeFamily: Cytochrome c oxidase subunit I-like3 c1fftF_
91.8
18
PDB header: oxidoreductaseChain: F: PDB Molecule: ubiquinol oxidase;PDBTitle: the structure of ubiquinol oxidase from escherichia coli
4 d1ar1a_
79.7
17
Fold: Cytochrome c oxidase subunit I-likeSuperfamily: Cytochrome c oxidase subunit I-likeFamily: Cytochrome c oxidase subunit I-like5 d3dtua1
76.8
18
Fold: Cytochrome c oxidase subunit I-likeSuperfamily: Cytochrome c oxidase subunit I-likeFamily: Cytochrome c oxidase subunit I-like6 c1m56G_
76.1
18
PDB header: oxidoreductaseChain: G: PDB Molecule: cytochrome c oxidase;PDBTitle: structure of cytochrome c oxidase from rhodobactor2 sphaeroides (wild type)
7 d1v54a_
75.7
15
Fold: Cytochrome c oxidase subunit I-likeSuperfamily: Cytochrome c oxidase subunit I-likeFamily: Cytochrome c oxidase subunit I-like8 c3o0rB_
40.0
10
PDB header: immune system/oxidoreductaseChain: B: PDB Molecule: nitric oxide reductase subunit b;PDBTitle: crystal structure of nitric oxide reductase from pseudomonas2 aeruginosa in complex with antibody fragment
9 d1xmea1
31.0
19
Fold: Cytochrome c oxidase subunit I-likeSuperfamily: Cytochrome c oxidase subunit I-likeFamily: Cytochrome c oxidase subunit I-like10 c3kp9A_
29.1
16
PDB header: blood coagulation,oxidoreductaseChain: A: PDB Molecule: vkorc1/thioredoxin domain protein;PDBTitle: structure of a bacterial homolog of vitamin k epoxide reductase
11 c2bbjB_
17.1
17
PDB header: metal transport/membrane proteinChain: B: PDB Molecule: divalent cation transport-related protein;PDBTitle: crystal structure of the cora mg2+ transporter
12 c3lw52_
16.2
21
PDB header: photosynthesisChain: 2: PDB Molecule: type ii chlorophyll a/b binding protein from photosystem i;PDB Fragment: residues 81-246;
PDBTitle: improved model of plant photosystem i
13 d2p3ha1
14.1
7
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like14 d2r2za1
13.5
36
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like15 d3deda1
11.5
36
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like16 d2o3ga1
11.0
29
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like17 c3fh6F_
10.3
8
PDB header: transport proteinChain: F: PDB Molecule: maltose transport system permease protein malf;PDBTitle: crystal structure of the resting state maltose transporter from e.2 coli
18 c3dedB_
9.5
36
PDB header: membrane proteinChain: B: PDB Molecule: probable hemolysin;PDBTitle: c-terminal domain of probable hemolysin from chromobacterium violaceum
19 c1hgvA_
9.1
23
PDB header: virusChain: A: PDB Molecule: ph75 inovirus major coat protein;PDBTitle: filamentous bacteriophage ph75
20 d2a65a1
8.2
15
Fold: SNF-likeSuperfamily: SNF-likeFamily: SNF-like21 d2gz4a1
not modelled
8.2
12
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: HD domain22 d2oaia1
not modelled
7.9
43
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like23 c3lemA_
not modelled
7.9
19
PDB header: hydrolaseChain: A: PDB Molecule: fructosyltransferase;PDBTitle: crystal structure of fructosyltransferase (d191a) from a. japonicus in2 complex with nystose
24 d2nqwa1
not modelled
7.5
21
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like25 c2l9uA_
not modelled
7.3
20
PDB header: membrane proteinChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-3;PDBTitle: spatial structure of dimeric erbb3 transmembrane domain
26 d2plsa1
not modelled
6.9
29
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like27 c3mepC_
not modelled
6.5
20
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: uncharacterized protein eca2234;PDBTitle: crystal structure of eca2234 protein from erwinia2 carotovora, northeast structural genomics consortium target3 ewr44
28 d2rk5a1
not modelled
6.5
36
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like29 c3kopB_
not modelled
6.4
22
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of protein with a cyclophilin-like fold2 (yp_831253.1) from arthrobacter sp. fb24 at 1.90 a resolution
30 c3k11A_
not modelled
6.0
13
PDB header: hydrolaseChain: A: PDB Molecule: putative glycosyl hydrolase;PDBTitle: crystal structure of putative glycosyl hydrolase (np_813087.1) from2 bacteroides thetaiotaomicron vpi-5482 at 1.80 a resolution
31 c3eh4A_
not modelled
5.9
13
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome c oxidase subunit 1;PDBTitle: structure of the reduced form of cytochrome ba3 oxidase from thermus2 thermophilus
32 c3llbA_
not modelled
5.8
36
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: the crystal structure of the protein pa3983 with unknown2 function from pseudomonas aeruginosa pao1
33 d2axta1
not modelled
5.5
18
Fold: Bacterial photosystem II reaction centre, L and M subunitsSuperfamily: Bacterial photosystem II reaction centre, L and M subunitsFamily: Bacterial photosystem II reaction centre, L and M subunits34 d2o1ra1
not modelled
5.1
14
Fold: FAD-binding/transporter-associated domain-likeSuperfamily: FAD-binding/transporter-associated domain-likeFamily: CorC/HlyC domain-like