1 c1j6qA_
82.3
17
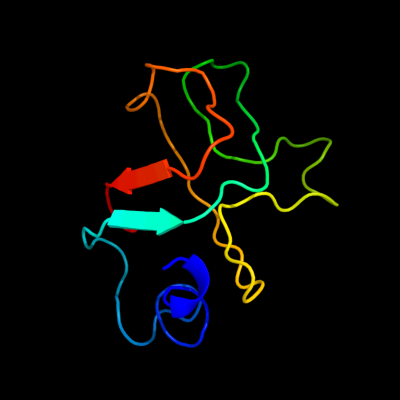
PDB header: chaperoneChain: A: PDB Molecule: cytochrome c maturation protein e;PDBTitle: solution structure and characterization of the heme2 chaperone ccme
2 d1j6qa_
82.3
17
Fold: OB-foldSuperfamily: Heme chaperone CcmEFamily: Heme chaperone CcmE3 d1sr3a_
66.0
21
Fold: OB-foldSuperfamily: Heme chaperone CcmEFamily: Heme chaperone CcmE4 c2kctA_
49.5
16
PDB header: chaperoneChain: A: PDB Molecule: cytochrome c-type biogenesis protein ccme;PDBTitle: solution nmr structure of the ob-fold domain of heme2 chaperone ccme from desulfovibrio vulgaris. northeast3 structural genomics target dvr115g.
5 c2r9qD_
40.9
22
PDB header: hydrolaseChain: D: PDB Molecule: 2'-deoxycytidine 5'-triphosphate deaminase;PDBTitle: crystal structure of 2'-deoxycytidine 5'-triphosphate deaminase from2 agrobacterium tumefaciens
6 c3h43F_
27.8
19
PDB header: hydrolaseChain: F: PDB Molecule: proteasome-activating nucleotidase;PDBTitle: n-terminal domain of the proteasome-activating nucleotidase2 of methanocaldococcus jannaschii
7 c2wkdA_
27.7
25
PDB header: dna binding proteinChain: A: PDB Molecule: orf34p2;PDBTitle: crystal structure of a double ile-to-met mutant of protein2 orf34 from lactococcus phage p2
8 d1n9wa1
26.4
16
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Anticodon-binding domain9 c2jwyA_
25.2
13
PDB header: lipoproteinChain: A: PDB Molecule: uncharacterized lipoprotein yaji;PDBTitle: solution nmr structure of uncharacterized lipoprotein yaji from2 escherichia coli. northeast structural genomics target er540
10 c2wg6L_
24.9
8
PDB header: transcription,hydrolaseChain: L: PDB Molecule: general control protein gcn4,PDBTitle: proteasome-activating nucleotidase (pan) n-domain (57-134)2 from archaeoglobus fulgidus fused to gcn4, p61a mutant
11 c2z14A_
24.6
17
PDB header: signaling proteinChain: A: PDB Molecule: ef-hand domain-containing family member c2;PDBTitle: crystal structure of the n-terminal duf1126 in human ef-2 hand domain containing 2 protein
12 c2wp8J_
23.5
28
PDB header: hydrolaseChain: J: PDB Molecule: exosome complex exonuclease dis3;PDBTitle: yeast rrp44 nuclease
13 d2pi2e1
20.2
15
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB14 d1l0wa1
18.1
18
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Anticodon-binding domain15 c1wydB_
17.9
12
PDB header: ligaseChain: B: PDB Molecule: hypothetical aspartyl-trna synthetase;PDBTitle: crystal structure of aspartyl-trna synthetase from sulfolobus tokodaii
16 d2vnud1
17.9
33
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like17 d1t3ta6
16.8
18
Fold: PurM C-terminal domain-likeSuperfamily: PurM C-terminal domain-likeFamily: PurM C-terminal domain-like18 c1vqwB_
14.4
14
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: protein with similarity to flavin-containingPDBTitle: crystal structure of a protein with similarity to flavin-2 containing monooxygenases and to mammalian dimethylalanine3 monooxygenases
19 c1yrlD_
11.9
20
PDB header: oxidoreductaseChain: D: PDB Molecule: ketol-acid reductoisomerase;PDBTitle: escherichia coli ketol-acid reductoisomerase
20 c3i7fA_
11.5
6
PDB header: ligaseChain: A: PDB Molecule: aspartyl-trna synthetase;PDBTitle: aspartyl trna synthetase from entamoeba histolytica
21 c2zauB_
not modelled
11.3
15
PDB header: transferaseChain: B: PDB Molecule: selenide, water dikinase;PDBTitle: crystal structure of an n-terminally truncated2 selenophosphate synthetase from aquifex aeolicus
22 d1ps9a2
not modelled
10.8
18
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: C-terminal domain of adrenodoxin reductase-like23 d2jn4a1
not modelled
10.3
35
Fold: NifT/FixU barrel-likeSuperfamily: NifT/FixU-likeFamily: NifT/FixU24 c2jn4A_
not modelled
10.3
35
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein fixu, nift;PDBTitle: solution nmr structure of protein rp4601 from2 rhodopseudomonas palustris. northeast structural genomics3 consortium target rpt2; ontario center for structural4 proteomics target rp4601.
25 d1so0a_
not modelled
9.7
19
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Aldose 1-epimerase (mutarotase)26 c2p39A_
not modelled
9.4
19
PDB header: signaling proteinChain: A: PDB Molecule: fibroblast growth factor 23;PDBTitle: crystal structure of human fgf23
27 c1pwaA_
not modelled
9.4
27
PDB header: hormone/growth factorChain: A: PDB Molecule: fibroblast growth factor-19;PDBTitle: crystal structure of fibroblast growth factor 19
28 d1pwaa_
not modelled
9.4
27
Fold: beta-TrefoilSuperfamily: CytokineFamily: Fibroblast growth factors (FGF)29 c2pqaB_
not modelled
9.2
13
PDB header: replicationChain: B: PDB Molecule: replication protein a 14 kda subunit;PDBTitle: crystal structure of full-length human rpa 14/32 heterodimer
30 c2zodB_
not modelled
9.1
15
PDB header: transferaseChain: B: PDB Molecule: selenide, water dikinase;PDBTitle: crystal structure of selenophosphate synthetase from2 aquifex aeolicus
31 c2pi2A_
not modelled
7.1
17
PDB header: replication, dna binding proteinChain: A: PDB Molecule: replication protein a 32 kda subunit;PDBTitle: full-length replication protein a subunits rpa14 and rpa32
32 d1geha2
not modelled
7.1
8
Fold: Ferredoxin-likeSuperfamily: RuBisCO, large subunit, small (N-terminal) domainFamily: Ribulose 1,5-bisphosphate carboxylase-oxygenase33 d1krta_
not modelled
7.0
8
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Anticodon-binding domain34 d1yloa1
not modelled
6.9
13
Fold: Domain of alpha and beta subunits of F1 ATP synthase-likeSuperfamily: Aminopeptidase/glucanase lid domainFamily: Aminopeptidase/glucanase lid domain35 d2z1ea2
not modelled
6.8
13
Fold: PurM C-terminal domain-likeSuperfamily: PurM C-terminal domain-likeFamily: PurM C-terminal domain-like36 d1c0aa1
not modelled
6.6
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Anticodon-binding domain37 c3m9bK_
not modelled
6.6
12
PDB header: chaperoneChain: K: PDB Molecule: proteasome-associated atpase;PDBTitle: crystal structure of the amino terminal coiled coil domain and the2 inter domain of the mycobacterium tuberculosis proteasomal atpase mpa
38 d2gycb1
not modelled
6.6
20
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Ribosomal protein L339 d2ijra1
not modelled
6.6
28
Fold: Api92-likeSuperfamily: Api92-likeFamily: Api92-like40 c1b8aB_
not modelled
6.4
22
PDB header: ligaseChain: B: PDB Molecule: protein (aspartyl-trna synthetase);PDBTitle: aspartyl-trna synthetase
41 c3stbC_
not modelled
6.2
9
PDB header: rna binding protein/immune systemChain: C: PDB Molecule: rna-editing complex protein mp42;PDBTitle: a complex of two editosome proteins and two nanobodies
42 d1b33n_
not modelled
6.1
50
Fold: Allophycocyanin linker chain (domain)Superfamily: Allophycocyanin linker chain (domain)Family: Allophycocyanin linker chain (domain)43 d2ns0a1
not modelled
6.0
32
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: RHA1 ro06458-like44 d2zoda2
not modelled
5.7
15
Fold: PurM C-terminal domain-likeSuperfamily: PurM C-terminal domain-likeFamily: PurM C-terminal domain-like45 c3gwdA_
not modelled
5.6
26
PDB header: oxidoreductaseChain: A: PDB Molecule: cyclohexanone monooxygenase;PDBTitle: closed crystal structure of cyclohexanone monooxygenase
46 d2d69a2
not modelled
5.6
20
Fold: Ferredoxin-likeSuperfamily: RuBisCO, large subunit, small (N-terminal) domainFamily: Ribulose 1,5-bisphosphate carboxylase-oxygenase47 d2j01e1
not modelled
5.4
20
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Ribosomal protein L348 c2esyA_
not modelled
5.4
45
PDB header: lipid binding proteinChain: A: PDB Molecule: lung surfactant protein c;PDBTitle: structure and influence on stability and activity of the n-2 terminal propetide part of lung surfactant protein c
49 d2gv8a2
not modelled
5.2
23
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains