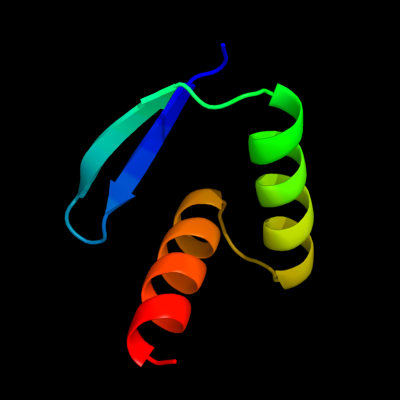
1 c1u9pA_
68.0
25
PDB header: unknown functionChain: A: PDB Molecule: parc;PDBTitle: permuted single-chain arc
2 c2vl7A_
39.4
21
PDB header: unknown functionChain: A: PDB Molecule: xpd;PDBTitle: structure of s. tokodaii xpd4
3 d1mnta_
32.8
25
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: Arc/Mnt-like phage repressors4 c2ehoL_
18.5
16
PDB header: replicationChain: L: PDB Molecule: gins complex subunit 3;PDBTitle: crystal structure of human gins complex
5 c1ezyA_
17.1
20
PDB header: signaling protein inhibitorChain: A: PDB Molecule: regulator of g-protein signaling 4;PDBTitle: high-resolution solution structure of free rgs4 by nmr
6 c3crw1_
16.9
19
PDB header: hydrolaseChain: 1: PDB Molecule: xpd/rad3 related dna helicase;PDBTitle: "xpd_apo"
7 d2jm5a1
15.6
22
Fold: Regulator of G-protein signaling, RGSSuperfamily: Regulator of G-protein signaling, RGSFamily: Regulator of G-protein signaling, RGS8 d3d85d1
15.0
42
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: I set domains9 c3pt2A_
14.3
4
PDB header: hydrolase/protein bindingChain: A: PDB Molecule: rna polymerase;PDBTitle: structure of a viral otu domain protease bound to ubiquitin
10 d1kxpd3
12.6
26
Fold: Serum albumin-likeSuperfamily: Serum albumin-likeFamily: Serum albumin-like11 c3c0rC_
12.2
16
PDB header: cell cycle, hydrolaseChain: C: PDB Molecule: ubiquitin thioesterase otu1;PDBTitle: structure of ovarian tumor (otu) domain in complex with ubiquitin
12 d1ufaa1
10.7
21
Fold: immunoglobulin/albumin-binding domain-likeSuperfamily: Families 57/38 glycoside transferase middle domainFamily: AmyC C-terminal domain-like13 c1ygmA_
10.4
29
PDB header: membrane proteinChain: A: PDB Molecule: hypothetical protein bsu31320;PDBTitle: nmr structure of mistic
14 d2b5dx1
10.4
21
Fold: immunoglobulin/albumin-binding domain-likeSuperfamily: Families 57/38 glycoside transferase middle domainFamily: AmyC C-terminal domain-like15 c3dtfB_
10.2
19
PDB header: transferaseChain: B: PDB Molecule: branched-chain amino acid aminotransferase;PDBTitle: structural analysis of mycobacterial branched chain aminotransferase-2 implications for inhibitor design
16 d1y6ia2
10.0
16
Fold: GUN4-likeSuperfamily: GUN4-likeFamily: GUN4-like17 c3cxbA_
9.8
25
PDB header: signaling proteinChain: A: PDB Molecule: protein sifa;PDBTitle: crystal structure of sifa and skip
18 d1914a2
9.1
17
Fold: Signal recognition particle alu RNA binding heterodimer, SRP9/14Superfamily: Signal recognition particle alu RNA binding heterodimer, SRP9/14Family: Signal recognition particle alu RNA binding heterodimer, SRP9/1419 d2ik8b1
8.3
24
Fold: Regulator of G-protein signaling, RGSSuperfamily: Regulator of G-protein signaling, RGSFamily: Regulator of G-protein signaling, RGS20 c2jpiA_
7.7
50
PDB header: structural genomicsChain: A: PDB Molecule: hypothetical protein;PDBTitle: chemical shift assignments of pa4090 from pseudomonas2 aeruginosa
21 c2abjG_
not modelled
7.5
10
PDB header: transferaseChain: G: PDB Molecule: branched-chain-amino-acid aminotransferase, cytosolic;PDBTitle: crystal structure of human branched chain amino acid transaminase in a2 complex with an inhibitor, c16h10n2o4f3scl, and pyridoxal 5'3 phosphate.
22 d2a1ha1
not modelled
7.4
10
Fold: D-aminoacid aminotransferase-like PLP-dependent enzymesSuperfamily: D-aminoacid aminotransferase-like PLP-dependent enzymesFamily: D-aminoacid aminotransferase-like PLP-dependent enzymes23 d1ljma_
not modelled
7.4
44
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: p53-like transcription factorsFamily: RUNT domain24 d1z3xa2
not modelled
6.9
13
Fold: GUN4-likeSuperfamily: GUN4-likeFamily: GUN4-like25 c3enpA_
not modelled
6.9
13
PDB header: hydrolaseChain: A: PDB Molecule: tp53rk-binding protein;PDBTitle: crystal structure of human cgi121
26 c3qqmD_
not modelled
6.8
25
PDB header: transferaseChain: D: PDB Molecule: mlr3007 protein;PDBTitle: crystal structure of a putative amino-acid aminotransferase2 (np_104211.1) from mesorhizobium loti at 2.30 a resolution
27 c3j0gO_
not modelled
6.3
30
PDB header: virusChain: O: PDB Molecule: e3 protein;PDBTitle: homology model of e3 protein of venezuelan equine encephalitis virus2 tc-83 strain fitted with a cryo-em map
28 d1eaqa_
not modelled
5.9
50
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: p53-like transcription factorsFamily: RUNT domain29 d1y9ba1
not modelled
5.7
26
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: VCA0319-like30 d1c4oa2
not modelled
5.7
29
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain31 c2konA_
not modelled
5.3
42
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: nmr solution structure of cv_2116 from chromobacterium2 violaceum. northeast structural genomics consortium target3 cvt4(1-82)
32 c2elxA_
not modelled
5.2
20
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 406;PDBTitle: solution structure of the 8th c2h2 zinc finger of mouse2 zinc finger protein 406