| 1 |
|
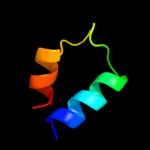
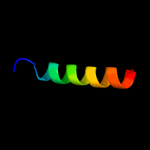
PDB 2rdc chain A
Region: 19 - 29
Aligned: 11
Modelled: 11
Confidence: 12.2%
Identity: 36%
PDB header:lipid binding protein
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a putative lipid binding protein (gsu0061) from2 geobacter sulfurreducens pca at 1.80 a resolution
Phyre2
| 2 |
|
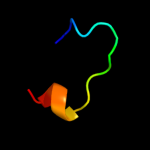
PDB 2ivx chain A domain 1
Region: 19 - 46
Aligned: 28
Modelled: 28
Confidence: 9.0%
Identity: 18%
Fold: Cyclin-like
Superfamily: Cyclin-like
Family: Cyclin
Phyre2
| 3 |
|
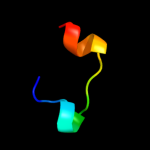
PDB 1j73 chain A
Region: 16 - 31
Aligned: 16
Modelled: 16
Confidence: 8.2%
Identity: 50%
PDB header:hormone/growth factor
Chain: A: PDB Molecule:insulin a;
PDBTitle: crystal structure of an unstable insulin analog with native activity.
Phyre2
| 4 |
|
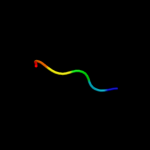
PDB 1j73 chain C
Region: 16 - 31
Aligned: 16
Modelled: 16
Confidence: 8.2%
Identity: 50%
PDB header:hormone/growth factor
Chain: C: PDB Molecule:insulin a;
PDBTitle: crystal structure of an unstable insulin analog with native activity.
Phyre2
| 5 |
|
PDB 3fq9 chain A
Region: 16 - 31
Aligned: 16
Modelled: 16
Confidence: 7.5%
Identity: 50%
PDB header:hormone
Chain: A: PDB Molecule:insulin;
PDBTitle: design of an insulin analog with enhanced receptor-binding2 selectivity. rationale, structure, and therapeutic3 implications
Phyre2
| 6 |
|
PDB 3fq9 chain C
Region: 16 - 31
Aligned: 16
Modelled: 16
Confidence: 7.5%
Identity: 50%
PDB header:hormone
Chain: C: PDB Molecule:insulin;
PDBTitle: design of an insulin analog with enhanced receptor-binding2 selectivity. rationale, structure, and therapeutic3 implications
Phyre2
| 7 |
|
PDB 3lem chain A
Region: 33 - 38
Aligned: 6
Modelled: 6
Confidence: 7.2%
Identity: 17%
PDB header:hydrolase
Chain: A: PDB Molecule:fructosyltransferase;
PDBTitle: crystal structure of fructosyltransferase (d191a) from a. japonicus in2 complex with nystose
Phyre2
| 8 |
|
PDB 3kf5 chain A
Region: 33 - 38
Aligned: 6
Modelled: 6
Confidence: 6.2%
Identity: 67%
PDB header:hydrolase
Chain: A: PDB Molecule:invertase;
PDBTitle: structure of invertase from schwanniomyces occidentalis
Phyre2
| 9 |
|
PDB 3ugf chain B
Region: 33 - 38
Aligned: 6
Modelled: 6
Confidence: 5.7%
Identity: 83%
PDB header:transferase
Chain: B: PDB Molecule:sucrose:(sucrose/fructan) 6-fructosyltransferase;
PDBTitle: crystal structure of a 6-sst/6-sft from pachysandra terminalis
Phyre2
| 10 |
|
PDB 3pij chain A
Region: 33 - 38
Aligned: 6
Modelled: 6
Confidence: 5.4%
Identity: 67%
PDB header:hydrolase
Chain: A: PDB Molecule:beta-fructofuranosidase;
PDBTitle: beta-fructofuranosidase from bifidobacterium longum - complex with2 fructose
Phyre2
| 11 |
|
PDB 3qz3 chain A
Region: 39 - 60
Aligned: 22
Modelled: 22
Confidence: 5.1%
Identity: 45%
PDB header:oxidoreductase
Chain: A: PDB Molecule:ferritin;
PDBTitle: the crystal structure of ferritin from vibrio cholerae o1 biovar el2 tor str. n16961
Phyre2