| 1 |
|
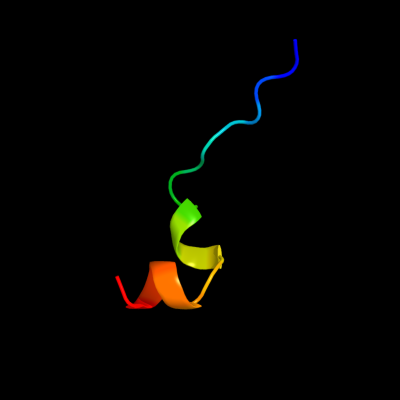
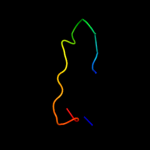
PDB 1kf6 chain C
Region: 68 - 87
Aligned: 20
Modelled: 20
Confidence: 20.4%
Identity: 20%
Fold: Heme-binding four-helical bundle
Superfamily: Fumarate reductase respiratory complex transmembrane subunits
Family: Succinate dehydrogenase/Fumarate reductase transmembrane subunits (SdhC/FrdC and SdhD/FrdD)
Phyre2
| 2 |
|
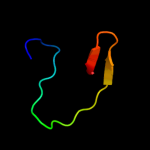
PDB 1bvp chain 1 domain 2
Region: 18 - 38
Aligned: 21
Modelled: 21
Confidence: 12.7%
Identity: 14%
Fold: Viral protein domain
Superfamily: Viral protein domain
Family: Top domain of virus capsid protein
Phyre2
| 3 |
|
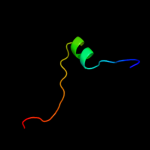
PDB 2de0 chain X
Region: 18 - 47
Aligned: 25
Modelled: 30
Confidence: 11.2%
Identity: 40%
PDB header:transferase
Chain: X: PDB Molecule:alpha-(1,6)-fucosyltransferase;
PDBTitle: crystal structure of human alpha 1,6-fucosyltransferase, fut8
Phyre2
| 4 |
|
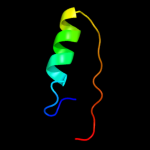
PDB 1ahs chain A
Region: 18 - 35
Aligned: 18
Modelled: 18
Confidence: 9.8%
Identity: 6%
Fold: Viral protein domain
Superfamily: Viral protein domain
Family: Top domain of virus capsid protein
Phyre2
| 5 |
|
PDB 3kwp chain A
Region: 4 - 31
Aligned: 28
Modelled: 28
Confidence: 9.6%
Identity: 7%
PDB header:transferase
Chain: A: PDB Molecule:predicted methyltransferase;
PDBTitle: crystal structure of putative methyltransferase from lactobacillus2 brevis
Phyre2
| 6 |
|
PDB 1wj2 chain A
Region: 45 - 50
Aligned: 6
Modelled: 6
Confidence: 9.4%
Identity: 67%
Fold: WRKY DNA-binding domain
Superfamily: WRKY DNA-binding domain
Family: WRKY DNA-binding domain
Phyre2
| 7 |
|
PDB 2pq4 chain B
Region: 10 - 30
Aligned: 21
Modelled: 21
Confidence: 9.2%
Identity: 29%
PDB header:chaperone/oxidoreductase
Chain: B: PDB Molecule:periplasmic nitrate reductase precursor;
PDBTitle: nmr solution structure of napd in complex with napa1-352 signal peptide
Phyre2
| 8 |
|
PDB 1h3g chain A domain 1
Region: 20 - 43
Aligned: 24
Modelled: 23
Confidence: 9.0%
Identity: 29%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: E set domains
Family: E-set domains of sugar-utilizing enzymes
Phyre2
| 9 |
|
PDB 2fpe chain B
Region: 18 - 44
Aligned: 27
Modelled: 27
Confidence: 8.5%
Identity: 15%
PDB header:signaling protein
Chain: B: PDB Molecule:c-jun-amino-terminal kinase interacting protein
PDBTitle: conserved dimerization of the ib1 src-homology 3 domain
Phyre2
| 10 |
|
PDB 2zom chain C
Region: 21 - 46
Aligned: 18
Modelled: 26
Confidence: 6.9%
Identity: 28%
PDB header:unknown function
Chain: C: PDB Molecule:protein cuta, chloroplast, putative, expressed;
PDBTitle: crystal structure of cuta1 from oryza sativa
Phyre2
| 11 |
|
PDB 2ayd chain A
Region: 45 - 50
Aligned: 6
Modelled: 6
Confidence: 6.8%
Identity: 67%
PDB header:transcription
Chain: A: PDB Molecule:wrky transcription factor 1;
PDBTitle: crystal structure of the c-terminal wrky domainof atwrky1,2 an sa-induced and partially npr1-dependent transcription3 factor
Phyre2
| 12 |
|
PDB 1s4d chain A
Region: 4 - 31
Aligned: 28
Modelled: 28
Confidence: 5.9%
Identity: 11%
Fold: Tetrapyrrole methylase
Superfamily: Tetrapyrrole methylase
Family: Tetrapyrrole methylase
Phyre2
| 13 |
|
PDB 3m3w chain A
Region: 41 - 48
Aligned: 8
Modelled: 8
Confidence: 5.5%
Identity: 25%
PDB header:endocytosis
Chain: A: PDB Molecule:protein kinase c and casein kinase ii substrate protein 3;
PDBTitle: crystal strcuture of mouse pacsin3 bar domain mutant
Phyre2