| 1 |
|
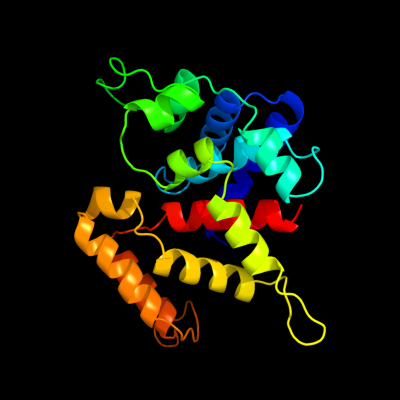
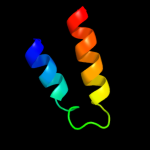
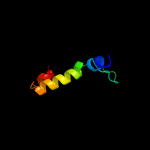
PDB 3h9p chain B
Region: 56 - 292
Aligned: 205
Modelled: 221
Confidence: 100.0%
Identity: 18%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative triphosphoribosyl-dephospho-coa synthase;
PDBTitle: crystal structure of putative triphosphoribosyl-dephospho-coa synthase2 from archaeoglobus fulgidus
Phyre2
| 2 |
|
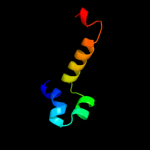
PDB 2xc7 chain A
Region: 228 - 279
Aligned: 40
Modelled: 52
Confidence: 28.5%
Identity: 28%
PDB header:rna binding protein
Chain: A: PDB Molecule:phosphorylated adapter rna export protein;
PDBTitle: solution structure of phax-rbd in complex with ssrna
Phyre2
| 3 |
|
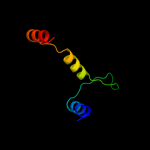
PDB 2w4s chain D
Region: 228 - 279
Aligned: 40
Modelled: 52
Confidence: 24.6%
Identity: 25%
PDB header:structural protein
Chain: D: PDB Molecule:ankyrin-repeat protein;
PDBTitle: novel rna-binding domain in cryptosporidium parvum at 2.52 angstrom resolution
Phyre2
| 4 |
|
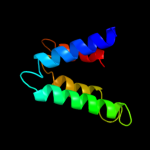
PDB 1p58 chain F
Region: 95 - 126
Aligned: 32
Modelled: 32
Confidence: 18.9%
Identity: 22%
PDB header:virus
Chain: F: PDB Molecule:envelope protein m;
PDBTitle: complex organization of dengue virus membrane proteins as revealed by2 9.5 angstrom cryo-em reconstruction
Phyre2
| 5 |
|
PDB 3c6a chain A
Region: 29 - 72
Aligned: 36
Modelled: 44
Confidence: 16.5%
Identity: 17%
PDB header:viral protein
Chain: A: PDB Molecule:terminase large subunit;
PDBTitle: crystal structure of the rb49 gp17 nuclease domain
Phyre2
| 6 |
|
PDB 3omd chain B
Region: 231 - 287
Aligned: 48
Modelled: 57
Confidence: 15.7%
Identity: 17%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of unknown function protein from leptospirillum2 rubarum
Phyre2
| 7 |
|
PDB 2f05 chain A domain 1
Region: 188 - 269
Aligned: 74
Modelled: 82
Confidence: 12.5%
Identity: 12%
Fold: PAH2 domain
Superfamily: PAH2 domain
Family: PAH2 domain
Phyre2
| 8 |
|
PDB 1w53 chain A
Region: 238 - 276
Aligned: 39
Modelled: 39
Confidence: 10.6%
Identity: 23%
Fold: KaiA/RbsU domain
Superfamily: KaiA/RbsU domain
Family: Phosphoserine phosphatase RsbU, N-terminal domain
Phyre2
| 9 |
|
PDB 1o22 chain A
Region: 260 - 277
Aligned: 18
Modelled: 18
Confidence: 9.8%
Identity: 22%
Fold: Hypothetical protein TM0875
Superfamily: Hypothetical protein TM0875
Family: Hypothetical protein TM0875
Phyre2
| 10 |
|
PDB 1q3q chain A domain 3
Region: 5 - 31
Aligned: 27
Modelled: 27
Confidence: 8.6%
Identity: 30%
Fold: GroEL-intermediate domain like
Superfamily: GroEL-intermediate domain like
Family: Group II chaperonin (CCT, TRIC), intermediate domain
Phyre2
| 11 |
|
PDB 1au7 chain A domain 2
Region: 258 - 277
Aligned: 20
Modelled: 20
Confidence: 7.6%
Identity: 30%
Fold: lambda repressor-like DNA-binding domains
Superfamily: lambda repressor-like DNA-binding domains
Family: POU-specific domain
Phyre2
| 12 |
|
PDB 1a6d chain B domain 3
Region: 5 - 31
Aligned: 27
Modelled: 27
Confidence: 7.3%
Identity: 22%
Fold: GroEL-intermediate domain like
Superfamily: GroEL-intermediate domain like
Family: Group II chaperonin (CCT, TRIC), intermediate domain
Phyre2
| 13 |
|
PDB 1a6d chain A domain 3
Region: 5 - 31
Aligned: 27
Modelled: 27
Confidence: 6.8%
Identity: 22%
Fold: GroEL-intermediate domain like
Superfamily: GroEL-intermediate domain like
Family: Group II chaperonin (CCT, TRIC), intermediate domain
Phyre2
| 14 |
|
PDB 1s5q chain B
Region: 188 - 269
Aligned: 74
Modelled: 82
Confidence: 6.2%
Identity: 12%
Fold: PAH2 domain
Superfamily: PAH2 domain
Family: PAH2 domain
Phyre2
| 15 |
|
PDB 2l9v chain A
Region: 260 - 279
Aligned: 20
Modelled: 20
Confidence: 6.1%
Identity: 15%
PDB header:rna binding protein
Chain: A: PDB Molecule:pre-mrna-processing factor 40 homolog a;
PDBTitle: nmr structure of the ff domain l24a mutant's folding transition state
Phyre2
| 16 |
|
PDB 1uou chain A domain 2
Region: 237 - 282
Aligned: 38
Modelled: 46
Confidence: 5.2%
Identity: 24%
Fold: Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Superfamily: Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Family: Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Phyre2