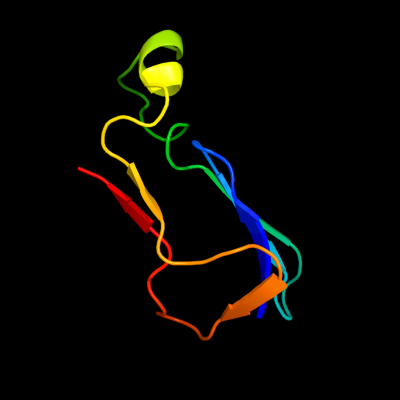
1 d1gpra_
62.6
20
Fold: Barrel-sandwich hybridSuperfamily: Duplicated hybrid motifFamily: Glucose permease-like2 d2f3ga_
58.5
18
Fold: Barrel-sandwich hybridSuperfamily: Duplicated hybrid motifFamily: Glucose permease-like3 d1u0ma2
56.5
9
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like4 d1glaf_
52.1
18
Fold: Barrel-sandwich hybridSuperfamily: Duplicated hybrid motifFamily: Glucose permease-like5 d2gpra_
38.1
19
Fold: Barrel-sandwich hybridSuperfamily: Duplicated hybrid motifFamily: Glucose permease-like6 d1pkla1
23.5
13
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: Pyruvate kinase beta-barrel domain7 c2dchX_
23.5
50
PDB header: hydrolaseChain: X: PDB Molecule: putative homing endonuclease;PDBTitle: crystal structure of archaeal intron-encoded homing endonuclease i-2 tsp061i
8 c2l55A_
20.8
20
PDB header: metal binding proteinChain: A: PDB Molecule: silb,silver efflux protein, mfp component of the threePDBTitle: solution structure of the c-terminal domain of silb from cupriavidus2 metallidurans
9 d1pkma1
18.7
20
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: Pyruvate kinase beta-barrel domain10 d1teda_
17.2
8
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like11 d1ycna_
15.3
17
Fold: AnnexinSuperfamily: AnnexinFamily: Annexin12 c1zeqX_
15.2
20
PDB header: metal binding proteinChain: X: PDB Molecule: cation efflux system protein cusf;PDBTitle: 1.5 a structure of apo-cusf residues 6-88 from escherichia2 coli
13 d1e0ta1
15.0
18
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: Pyruvate kinase beta-barrel domain14 d1apja_
13.6
26
Fold: TB module/8-cys domainSuperfamily: TB module/8-cys domainFamily: TB module/8-cys domain15 d2g50a1
13.1
20
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: Pyruvate kinase beta-barrel domain16 d1liua1
12.9
11
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: Pyruvate kinase beta-barrel domain17 c1u0mA_
12.7
8
PDB header: biosynthetic proteinChain: A: PDB Molecule: putative polyketide synthase;PDBTitle: crystal structure of 1,3,6,8-tetrahydroxynaphthalene synthase (thns)2 from streptomyces coelicolor a3(2): a bacterial type iii polyketide3 synthase (pks) provides insights into enzymatic control of reactive4 polyketide intermediates
18 c3aleB_
11.3
12
PDB header: transferaseChain: B: PDB Molecule: os07g0271500 protein;PDBTitle: a type iii polyketide synthase that produces diarylheptanoid
19 d1ksqa_
11.0
22
Fold: TB module/8-cys domainSuperfamily: TB module/8-cys domainFamily: TB module/8-cys domain20 c2d1kC_
10.5
36
PDB header: structural proteinChain: C: PDB Molecule: metastasis suppressor protein 1;PDBTitle: ternary complex of the wh2 domain of mim with actin-dnase i
21 c3oitB_
not modelled
10.1
13
PDB header: transferaseChain: B: PDB Molecule: os07g0271500 protein;PDBTitle: crystal structure of curcuminoid synthase cus from oryza sativa
22 c3ov3A_
not modelled
9.4
17
PDB header: transferaseChain: A: PDB Molecule: curcumin synthase;PDBTitle: g211f mutant of curcumin synthase 1 from curcuma longa
23 d1a3xa1
not modelled
9.1
9
Fold: PK beta-barrel domain-likeSuperfamily: PK beta-barrel domain-likeFamily: Pyruvate kinase beta-barrel domain24 c2p0uB_
not modelled
9.0
10
PDB header: transferaseChain: B: PDB Molecule: stilbenecarboxylate synthase 2;PDBTitle: crystal structure of marchantia polymorpha stilbenecarboxylate2 synthase 2 (stcs2)
25 c2e28A_
not modelled
8.7
20
PDB header: transferaseChain: A: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure analysis of pyruvate kinase from bacillus2 stearothermophilus
26 d1u0ua2
not modelled
8.6
11
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like27 c3mmlE_
not modelled
8.3
18
PDB header: hydrolaseChain: E: PDB Molecule: allophanate hydrolase subunit 2;PDBTitle: allophanate hydrolase complex from mycobacterium smegmatis, msmeg0435-2 msmeg0436
28 c3u1nC_
not modelled
8.1
19
PDB header: hydrolaseChain: C: PDB Molecule: sam domain and hd domain-containing protein 1;PDBTitle: structure of the catalytic core of human samhd1
29 c3a5qA_
not modelled
7.6
13
PDB header: transferaseChain: A: PDB Molecule: benzalacetone synthase;PDBTitle: benzalacetone synthase from rheum palmatum
30 d1fasa_
not modelled
7.1
57
Fold: Snake toxin-likeSuperfamily: Snake toxin-likeFamily: Snake venom toxins31 c3t07D_
not modelled
7.0
15
PDB header: transferase/transferase inhibitorChain: D: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure of s. aureus pyruvate kinase in complex with a2 naturally occurring bis-indole alkaloid
32 c2h84A_
not modelled
7.0
15
PDB header: biosynthetic protein, transferaseChain: A: PDB Molecule: steely1;PDBTitle: crystal structure of the c-terminal type iii polyketide synthase (pks2 iii) domain of 'steely1' (a type i/iii pks hybrid from dictyostelium)
33 d1m98a1
not modelled
6.9
9
Fold: Orange carotenoid protein, N-terminal domainSuperfamily: Orange carotenoid protein, N-terminal domainFamily: Orange carotenoid protein, N-terminal domain34 d1ve2a1
not modelled
6.8
47
Fold: Tetrapyrrole methylaseSuperfamily: Tetrapyrrole methylaseFamily: Tetrapyrrole methylase35 d1twda_
not modelled
6.7
100
Fold: TIM beta/alpha-barrelSuperfamily: CutC-likeFamily: CutC-like36 c1ee0A_
not modelled
6.5
6
PDB header: transferaseChain: A: PDB Molecule: 2-pyrone synthase;PDBTitle: 2-pyrone synthase complexed with acetoacetyl-coa
37 d1cnza_
not modelled
6.5
33
Fold: Isocitrate/Isopropylmalate dehydrogenase-likeSuperfamily: Isocitrate/Isopropylmalate dehydrogenase-likeFamily: Dimeric isocitrate & isopropylmalate dehydrogenases38 c3iwpK_
not modelled
6.4
80
PDB header: metal binding proteinChain: K: PDB Molecule: copper homeostasis protein cutc homolog;PDBTitle: crystal structure of human copper homeostasis protein cutc
39 c2q14A_
not modelled
6.4
19
PDB header: hydrolaseChain: A: PDB Molecule: phosphohydrolase;PDBTitle: crystal structure of phosphohydrolase (bt4208) from bacteroides2 thetaiotaomicron vpi-5482 at 2.20 a resolution
40 c1m98A_
not modelled
6.3
9
PDB header: unknown functionChain: A: PDB Molecule: orange carotenoid protein;PDBTitle: crystal structure of orange carotenoid protein
41 d1h9ma1
not modelled
5.9
22
Fold: OB-foldSuperfamily: MOP-likeFamily: BiMOP, duplicated molybdate-binding domain42 c2e0kA_
not modelled
5.9
41
PDB header: transferaseChain: A: PDB Molecule: precorrin-2 c20-methyltransferase;PDBTitle: crystal structure of cbil, a methyltransferase involved in anaerobic2 vitamin b12 biosynthesis
43 d1guta_
not modelled
5.9
14
Fold: OB-foldSuperfamily: MOP-likeFamily: Molybdate/tungstate binding protein MOP44 c3nutC_
not modelled
5.5
41
PDB header: transferaseChain: C: PDB Molecule: precorrin-3 methylase;PDBTitle: crystal structure of the methyltransferase cobj
45 c3eoeC_
not modelled
5.5
12
PDB header: transferaseChain: C: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure of pyruvate kinase from toxoplasma gondii, 55.m00007
46 d1drsa_
not modelled
5.5
38
Fold: Snake toxin-likeSuperfamily: Snake toxin-likeFamily: Dendroaspin47 c2bb3B_
not modelled
5.3
22
PDB header: transferaseChain: B: PDB Molecule: cobalamin biosynthesis precorrin-6y methylase (cbie);PDBTitle: crystal structure of cobalamin biosynthesis precorrin-6y methylase2 (cbie) from archaeoglobus fulgidus
48 c2j5dA_
not modelled
5.2
21
PDB header: membrane proteinChain: A: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: nmr structure of bnip3 transmembrane domain in lipid2 bicelles