| 1 |
|
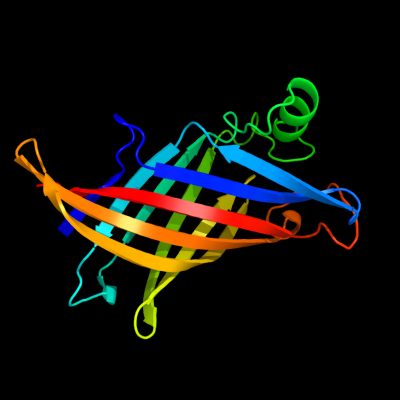
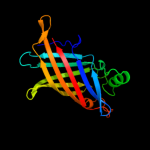
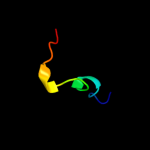
PDB 1y0g chain A
Region: 23 - 191
Aligned: 169
Modelled: 169
Confidence: 100.0%
Identity: 100%
Fold: Streptavidin-like
Superfamily: YceI-like
Family: YceI-like
Phyre2
| 2 |
|
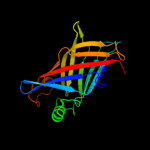
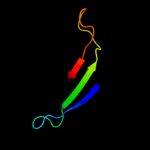
PDB 2fgs chain A
Region: 24 - 191
Aligned: 166
Modelled: 168
Confidence: 100.0%
Identity: 37%
PDB header:lipid binding protein
Chain: A: PDB Molecule:putative periplasmic protein;
PDBTitle: crystal structure of campylobacter jejuni ycei protein,2 structural genomics
Phyre2
| 3 |
|
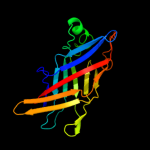
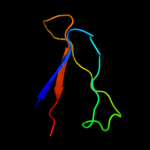
PDB 1wub chain A
Region: 24 - 190
Aligned: 164
Modelled: 167
Confidence: 100.0%
Identity: 35%
Fold: Streptavidin-like
Superfamily: YceI-like
Family: YceI-like
Phyre2
| 4 |
|
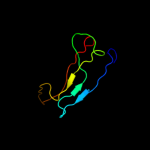
PDB 3hpe chain B
Region: 23 - 188
Aligned: 160
Modelled: 166
Confidence: 100.0%
Identity: 36%
PDB header:transport protein
Chain: B: PDB Molecule:conserved hypothetical secreted protein;
PDBTitle: crystal structure of ycei (hp1286) from helicobacter pylori
Phyre2
| 5 |
|
PDB 3q34 chain A
Region: 23 - 191
Aligned: 157
Modelled: 169
Confidence: 100.0%
Identity: 17%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ycei-like family protein;
PDBTitle: the crystal structure of ycei-like family protein from pseudomonas2 syringae
Phyre2
| 6 |
|
PDB 2x34 chain A
Region: 24 - 190
Aligned: 155
Modelled: 167
Confidence: 100.0%
Identity: 21%
PDB header:carbohydrate-binding protein
Chain: A: PDB Molecule:cellulose-binding protein, x158;
PDBTitle: structure of a polyisoprenoid binding domain from2 saccharophagus degradans implicated in plant cell wall3 breakdown
Phyre2
| 7 |
|
PDB 3uc0 chain B
Region: 99 - 167
Aligned: 54
Modelled: 69
Confidence: 29.3%
Identity: 20%
PDB header:viral protein/immune system
Chain: B: PDB Molecule:envelope protein;
PDBTitle: crystal structure of domain i of the envelope glycoprotein ectodomain2 from dengue virus serotype 4 in complex with the fab fragment of the3 chimpanzee monoclonal antibody 5h2
Phyre2
| 8 |
|
PDB 2ich chain A domain 1
Region: 119 - 135
Aligned: 17
Modelled: 17
Confidence: 9.6%
Identity: 35%
Fold: AttH-like
Superfamily: AttH-like
Family: AttH-like
Phyre2
| 9 |
|
PDB 2cg9 chain A
Region: 165 - 182
Aligned: 18
Modelled: 18
Confidence: 8.7%
Identity: 17%
PDB header:chaperone
Chain: A: PDB Molecule:atp-dependent molecular chaperone hsp82;
PDBTitle: crystal structure of an hsp90-sba1 closed chaperone complex
Phyre2
| 10 |
|
PDB 2o39 chain A domain 1
Region: 121 - 156
Aligned: 36
Modelled: 36
Confidence: 7.8%
Identity: 11%
Fold: Virus attachment protein globular domain
Superfamily: Virus attachment protein globular domain
Family: Adenovirus fiber protein "knob" domain
Phyre2
| 11 |
|
PDB 2qlv chain B domain 1
Region: 35 - 78
Aligned: 44
Modelled: 44
Confidence: 5.6%
Identity: 16%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: E set domains
Family: AMPK-beta glycogen binding domain-like
Phyre2