| 1 |
|
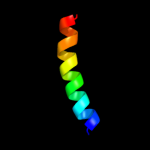
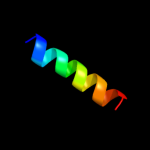
PDB 2gmq chain A domain 1
Region: 128 - 157
Aligned: 29
Modelled: 30
Confidence: 16.3%
Identity: 10%
Fold: PUA domain-like
Superfamily: PUA domain-like
Family: PrgU-like
Phyre2
| 2 |
|
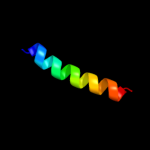
PDB 8tfv chain A
Region: 149 - 160
Aligned: 12
Modelled: 12
Confidence: 14.6%
Identity: 33%
PDB header:antimicrobial
Chain: A: PDB Molecule:protein (thanatin);
PDBTitle: insect defense peptide
Phyre2
| 3 |
|
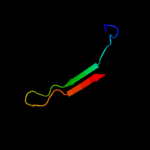
PDB 1cid chain A domain 1
Region: 42 - 91
Aligned: 46
Modelled: 50
Confidence: 13.4%
Identity: 13%
Fold: Immunoglobulin-like beta-sandwich
Superfamily: Immunoglobulin
Family: V set domains (antibody variable domain-like)
Phyre2
| 4 |
|
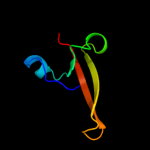
PDB 1q6w chain A
Region: 32 - 74
Aligned: 43
Modelled: 43
Confidence: 11.3%
Identity: 16%
Fold: Thioesterase/thiol ester dehydrase-isomerase
Superfamily: Thioesterase/thiol ester dehydrase-isomerase
Family: MaoC-like
Phyre2
| 5 |
|
PDB 1cid chain A
Region: 42 - 115
Aligned: 69
Modelled: 74
Confidence: 10.6%
Identity: 14%
PDB header:t-cell surface glycoprotein
Chain: A: PDB Molecule:t cell surface glycoprotein cd4;
PDBTitle: crystal structure of domains 3 & 4 of rat cd4 and their2 relationship to the nh2-terminal domains
Phyre2
| 6 |
|
PDB 1ppj chain D domain 2
Region: 3 - 24
Aligned: 22
Modelled: 22
Confidence: 9.7%
Identity: 27%
Fold: Single transmembrane helix
Superfamily: Cytochrome c1 subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase), transmembrane anchor
Family: Cytochrome c1 subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase), transmembrane anchor
Phyre2
| 7 |
|
PDB 1azp chain A
Region: 78 - 95
Aligned: 18
Modelled: 18
Confidence: 8.6%
Identity: 11%
Fold: SH3-like barrel
Superfamily: Chromo domain-like
Family: "Histone-like" proteins from archaea
Phyre2
| 8 |
|
PDB 3cx5 chain D domain 2
Region: 3 - 24
Aligned: 22
Modelled: 22
Confidence: 7.9%
Identity: 23%
Fold: Single transmembrane helix
Superfamily: Cytochrome c1 subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase), transmembrane anchor
Family: Cytochrome c1 subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase), transmembrane anchor
Phyre2
| 9 |
|
PDB 3if8 chain A
Region: 151 - 160
Aligned: 10
Modelled: 10
Confidence: 6.2%
Identity: 10%
PDB header:cell cycle
Chain: A: PDB Molecule:protein zwilch homolog;
PDBTitle: crystal structure of zwilch, a member of the rzz kinetochore complex
Phyre2
| 10 |
|
PDB 2bo9 chain B
Region: 30 - 57
Aligned: 28
Modelled: 28
Confidence: 5.7%
Identity: 14%
PDB header:hydrolase
Chain: B: PDB Molecule:human latexin;
PDBTitle: human carboxypeptidase a4 in complex with human latexin.
Phyre2
| 11 |
|
PDB 1qcs chain A domain 1
Region: 101 - 154
Aligned: 50
Modelled: 51
Confidence: 5.7%
Identity: 12%
Fold: Double psi beta-barrel
Superfamily: ADC-like
Family: Cdc48 N-terminal domain-like
Phyre2
| 12 |
|
PDB 3arc chain T
Region: 8 - 25
Aligned: 18
Modelled: 18
Confidence: 5.6%
Identity: 22%
PDB header:electron transport, photosynthesis
Chain: T: PDB Molecule:photosystem ii reaction center protein t;
PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
Phyre2
| 13 |
|
PDB 3a0b chain T
Region: 8 - 25
Aligned: 18
Modelled: 18
Confidence: 5.6%
Identity: 22%
PDB header:electron transport
Chain: T: PDB Molecule:photosystem ii reaction center protein t;
PDBTitle: crystal structure of br-substituted photosystem ii complex
Phyre2
| 14 |
|
PDB 2j7a chain C
Region: 9 - 37
Aligned: 29
Modelled: 29
Confidence: 5.1%
Identity: 10%
PDB header:oxidoreductase
Chain: C: PDB Molecule:cytochrome c quinol dehydrogenase nrfh;
PDBTitle: crystal structure of cytochrome c nitrite reductase nrfha2 complex from desulfovibrio vulgaris
Phyre2