| 1 |
|
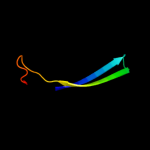
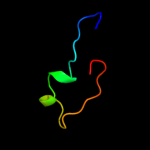
PDB 3hm0 chain C
Region: 11 - 42
Aligned: 32
Modelled: 32
Confidence: 13.0%
Identity: 13%
PDB header:hydrolase
Chain: C: PDB Molecule:probable thioesterase;
PDBTitle: crystal structure of probable thioesterase from bartonella2 henselae
Phyre2
| 2 |
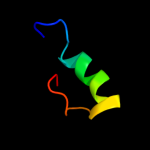
|
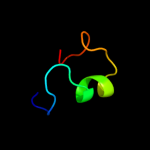
PDB 2xs4 chain A
Region: 9 - 34
Aligned: 16
Modelled: 26
Confidence: 12.1%
Identity: 31%
PDB header:hydrolase
Chain: A: PDB Molecule:karilysin protease;
PDBTitle: structure of karilysin catalytic mmp domain in complex with2 magnesium
Phyre2
| 3 |
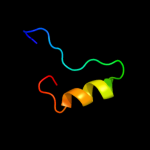
|
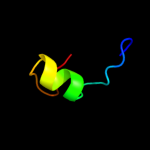
PDB 1eak chain A domain 2
Region: 9 - 34
Aligned: 16
Modelled: 26
Confidence: 9.5%
Identity: 31%
Fold: Zincin-like
Superfamily: Metalloproteases ("zincins"), catalytic domain
Family: Matrix metalloproteases, catalytic domain
Phyre2
| 4 |
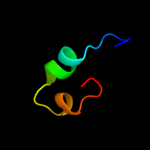
|
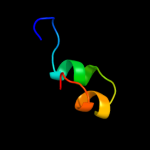
PDB 1xuc chain A domain 1
Region: 9 - 34
Aligned: 16
Modelled: 26
Confidence: 9.0%
Identity: 44%
Fold: Zincin-like
Superfamily: Metalloproteases ("zincins"), catalytic domain
Family: Matrix metalloproteases, catalytic domain
Phyre2
| 5 |
|
PDB 1hov chain A
Region: 9 - 34
Aligned: 16
Modelled: 26
Confidence: 8.8%
Identity: 31%
Fold: Zincin-like
Superfamily: Metalloproteases ("zincins"), catalytic domain
Family: Matrix metalloproteases, catalytic domain
Phyre2
| 6 |
|
PDB 1hfc chain A
Region: 9 - 34
Aligned: 16
Modelled: 26
Confidence: 8.7%
Identity: 38%
Fold: Zincin-like
Superfamily: Metalloproteases ("zincins"), catalytic domain
Family: Matrix metalloproteases, catalytic domain
Phyre2
| 7 |
|
PDB 1cxv chain A
Region: 9 - 34
Aligned: 16
Modelled: 26
Confidence: 8.5%
Identity: 44%
Fold: Zincin-like
Superfamily: Metalloproteases ("zincins"), catalytic domain
Family: Matrix metalloproteases, catalytic domain
Phyre2
| 8 |
|
PDB 1mmq chain A
Region: 9 - 32
Aligned: 14
Modelled: 24
Confidence: 8.5%
Identity: 43%
Fold: Zincin-like
Superfamily: Metalloproteases ("zincins"), catalytic domain
Family: Matrix metalloproteases, catalytic domain
Phyre2
| 9 |
|
PDB 1fbl chain A domain 2
Region: 9 - 34
Aligned: 16
Modelled: 26
Confidence: 8.4%
Identity: 38%
Fold: Zincin-like
Superfamily: Metalloproteases ("zincins"), catalytic domain
Family: Matrix metalloproteases, catalytic domain
Phyre2
| 10 |
|
PDB 1q3a chain A
Region: 9 - 34
Aligned: 16
Modelled: 26
Confidence: 8.1%
Identity: 31%
Fold: Zincin-like
Superfamily: Metalloproteases ("zincins"), catalytic domain
Family: Matrix metalloproteases, catalytic domain
Phyre2
| 11 |
|
PDB 1hy7 chain A
Region: 9 - 34
Aligned: 16
Modelled: 26
Confidence: 7.9%
Identity: 31%
Fold: Zincin-like
Superfamily: Metalloproteases ("zincins"), catalytic domain
Family: Matrix metalloproteases, catalytic domain
Phyre2
| 12 |
|
PDB 1hv5 chain A
Region: 9 - 34
Aligned: 16
Modelled: 26
Confidence: 7.2%
Identity: 31%
Fold: Zincin-like
Superfamily: Metalloproteases ("zincins"), catalytic domain
Family: Matrix metalloproteases, catalytic domain
Phyre2
| 13 |
|
PDB 1rm8 chain A
Region: 9 - 15
Aligned: 7
Modelled: 7
Confidence: 6.2%
Identity: 71%
Fold: Zincin-like
Superfamily: Metalloproteases ("zincins"), catalytic domain
Family: Matrix metalloproteases, catalytic domain
Phyre2
| 14 |
|
PDB 3bbn chain M
Region: 1 - 13
Aligned: 13
Modelled: 13
Confidence: 5.5%
Identity: 46%
PDB header:ribosome
Chain: M: PDB Molecule:ribosomal protein s13;
PDBTitle: homology model for the spinach chloroplast 30s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome.
Phyre2
| 15 |
|
PDB 1y93 chain A domain 1
Region: 9 - 15
Aligned: 7
Modelled: 7
Confidence: 5.3%
Identity: 71%
Fold: Zincin-like
Superfamily: Metalloproteases ("zincins"), catalytic domain
Family: Matrix metalloproteases, catalytic domain
Phyre2
| 16 |
|
PDB 2ovx chain A domain 1
Region: 9 - 15
Aligned: 7
Modelled: 7
Confidence: 5.2%
Identity: 86%
Fold: Zincin-like
Superfamily: Metalloproteases ("zincins"), catalytic domain
Family: Matrix metalloproteases, catalytic domain
Phyre2
| 17 |
|
PDB 2hu9 chain B
Region: 33 - 39
Aligned: 7
Modelled: 7
Confidence: 5.0%
Identity: 71%
PDB header:metal transport
Chain: B: PDB Molecule:mercuric transport protein periplasmic component;
PDBTitle: x-ray structure of the archaeoglobus fulgidus copz n-2 terminal domain
Phyre2