| 1 |
|
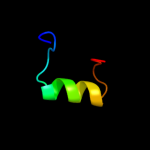
PDB 2oar chain A
Region: 142 - 195
Aligned: 54
Modelled: 54
Confidence: 24.9%
Identity: 13%
PDB header:membrane protein
Chain: A: PDB Molecule:large-conductance mechanosensitive channel;
PDBTitle: mechanosensitive channel of large conductance (mscl)
Phyre2
| 2 |
|
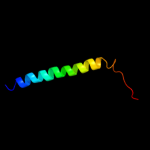
PDB 1ppj chain C domain 2
Region: 180 - 248
Aligned: 69
Modelled: 69
Confidence: 18.4%
Identity: 13%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 3 |
|
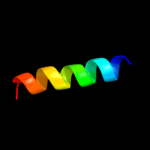
PDB 3rlb chain A
Region: 223 - 341
Aligned: 109
Modelled: 119
Confidence: 18.1%
Identity: 26%
PDB header:thiamine-binding protein
Chain: A: PDB Molecule:thit;
PDBTitle: crystal structure at 2.0 a of the s-component for thiamin from an ecf-2 type abc transporter
Phyre2
| 4 |
|
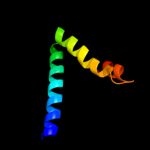
PDB 1bcc chain C domain 3
Region: 180 - 247
Aligned: 68
Modelled: 68
Confidence: 11.4%
Identity: 13%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 5 |
|
PDB 1kkx chain A
Region: 281 - 345
Aligned: 64
Modelled: 65
Confidence: 10.7%
Identity: 6%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: ARID-like
Family: ARID domain
Phyre2
| 6 |
|
PDB 3cwb chain C
Region: 180 - 248
Aligned: 69
Modelled: 69
Confidence: 10.2%
Identity: 13%
PDB header:oxidoreductase
Chain: C: PDB Molecule:cytochrome b;
PDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d
Phyre2
| 7 |
|
PDB 2oar chain A domain 1
Region: 142 - 189
Aligned: 48
Modelled: 48
Confidence: 10.2%
Identity: 15%
Fold: Gated mechanosensitive channel
Superfamily: Gated mechanosensitive channel
Family: Gated mechanosensitive channel
Phyre2
| 8 |
|
PDB 2r6g chain F domain 1
Region: 12 - 89
Aligned: 78
Modelled: 78
Confidence: 7.9%
Identity: 12%
Fold: MalF N-terminal region-like
Superfamily: MalF N-terminal region-like
Family: MalF N-terminal region-like
Phyre2
| 9 |
|
PDB 1r11 chain A domain 3
Region: 285 - 307
Aligned: 23
Modelled: 23
Confidence: 6.0%
Identity: 13%
Fold: MutS N-terminal domain-like
Superfamily: tRNA-intron endonuclease N-terminal domain-like
Family: tRNA-intron endonuclease N-terminal domain-like
Phyre2
| 10 |
|
PDB 3ixz chain B
Region: 244 - 286
Aligned: 41
Modelled: 41
Confidence: 5.9%
Identity: 12%
PDB header:hydrolase
Chain: B: PDB Molecule:potassium-transporting atpase subunit beta;
PDBTitle: pig gastric h+/k+-atpase complexed with aluminium fluoride
Phyre2
| 11 |
|
PDB 2axt chain J domain 1
Region: 296 - 313
Aligned: 18
Modelled: 18
Confidence: 5.9%
Identity: 11%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction center protein J, PsbJ
Family: PsbJ-like
Phyre2
| 12 |
|
PDB 3cx5 chain C domain 2
Region: 195 - 248
Aligned: 54
Modelled: 54
Confidence: 5.7%
Identity: 13%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2
| 13 |
|
PDB 2qjk chain M
Region: 195 - 248
Aligned: 54
Modelled: 54
Confidence: 5.5%
Identity: 13%
PDB header:electron transport
Chain: M: PDB Molecule:cytochrome b;
PDBTitle: crystal structure analysis of mutant rhodobacter2 sphaeroides bc1 with stigmatellin and antimycin
Phyre2
| 14 |
|
PDB 2e74 chain A domain 1
Region: 195 - 248
Aligned: 54
Modelled: 54
Confidence: 5.3%
Identity: 11%
Fold: Heme-binding four-helical bundle
Superfamily: Transmembrane di-heme cytochromes
Family: Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
Phyre2