| 1 |
|
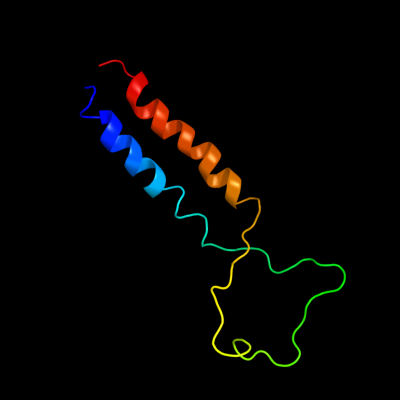
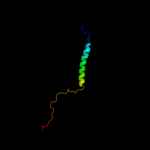
PDB 2aki chain A
Region: 1 - 77
Aligned: 77
Modelled: 77
Confidence: 100.0%
Identity: 100%
PDB header:protein transport
Chain: A: PDB Molecule:protein-export membrane protein secg;
PDBTitle: normal mode-based flexible fitted coordinates of a2 translocating secyeg protein-conducting channel into the3 cryo-em map of a secyeg-nascent chain-70s ribosome complex4 from e. coli
Phyre2
| 2 |
|
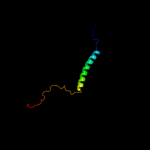
PDB 3din chain E
Region: 9 - 73
Aligned: 65
Modelled: 65
Confidence: 99.9%
Identity: 26%
PDB header:membrane protein, protein transport
Chain: E: PDB Molecule:preprotein translocase subunit secg;
PDBTitle: crystal structure of the protein-translocation complex formed by the2 secy channel and the seca atpase
Phyre2
| 3 |
|
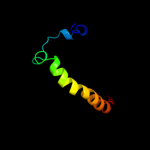
PDB 3dl8 chain F
Region: 8 - 73
Aligned: 65
Modelled: 66
Confidence: 99.9%
Identity: 38%
PDB header:protein transport
Chain: F: PDB Molecule:protein-export membrane protein secg;
PDBTitle: structure of the complex of aquifex aeolicus secyeg and2 bacillus subtilis seca
Phyre2
| 4 |
|
PDB 2axt chain I domain 1
Region: 54 - 82
Aligned: 29
Modelled: 29
Confidence: 56.2%
Identity: 17%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction center protein I, PsbI
Family: PsbI-like
Phyre2
| 5 |
|
PDB 2y69 chain W
Region: 42 - 78
Aligned: 37
Modelled: 37
Confidence: 21.0%
Identity: 19%
PDB header:electron transport
Chain: W: PDB Molecule:cytochrome c oxidase polypeptide 7a1;
PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
Phyre2
| 6 |
|
PDB 1m56 chain D
Region: 46 - 75
Aligned: 29
Modelled: 30
Confidence: 18.6%
Identity: 14%
Fold: Single transmembrane helix
Superfamily: Bacterial aa3 type cytochrome c oxidase subunit IV
Family: Bacterial aa3 type cytochrome c oxidase subunit IV
Phyre2
| 7 |
|
PDB 1zll chain E
Region: 4 - 22
Aligned: 19
Modelled: 19
Confidence: 16.1%
Identity: 11%
PDB header:membrane protein/signaling protein
Chain: E: PDB Molecule:cardiac phospholamban;
PDBTitle: nmr structure of unphosphorylated human phospholamban2 pentamer
Phyre2
| 8 |
|
PDB 1v54 chain J
Region: 42 - 79
Aligned: 38
Modelled: 38
Confidence: 10.4%
Identity: 18%
Fold: Single transmembrane helix
Superfamily: Mitochondrial cytochrome c oxidase subunit VIIa
Family: Mitochondrial cytochrome c oxidase subunit VIIa
Phyre2
| 9 |
|
PDB 1z65 chain A
Region: 56 - 79
Aligned: 24
Modelled: 24
Confidence: 10.3%
Identity: 17%
PDB header:unknown function
Chain: A: PDB Molecule:prion-like protein doppel;
PDBTitle: mouse doppel 1-30 peptide
Phyre2
| 10 |
|
PDB 1yew chain I
Region: 45 - 105
Aligned: 61
Modelled: 61
Confidence: 10.2%
Identity: 10%
PDB header:oxidoreductase, membrane protein
Chain: I: PDB Molecule:particulate methane monooxygenase, b subunit;
PDBTitle: crystal structure of particulate methane monooxygenase
Phyre2
| 11 |
|
PDB 3rgb chain A
Region: 45 - 105
Aligned: 61
Modelled: 61
Confidence: 10.2%
Identity: 10%
PDB header:oxidoreductase
Chain: A: PDB Molecule:methane monooxygenase subunit b2;
PDBTitle: crystal structure of particulate methane monooxygenase from2 methylococcus capsulatus (bath)
Phyre2
| 12 |
|
PDB 1a62 chain A domain 1
Region: 16 - 29
Aligned: 14
Modelled: 14
Confidence: 9.3%
Identity: 21%
Fold: LEM/SAP HeH motif
Superfamily: Rho N-terminal domain-like
Family: Rho termination factor, N-terminal domain
Phyre2
| 13 |
|
PDB 2k21 chain A
Region: 30 - 83
Aligned: 54
Modelled: 54
Confidence: 6.2%
Identity: 19%
PDB header:membrane protein
Chain: A: PDB Molecule:potassium voltage-gated channel subfamily e
PDBTitle: nmr structure of human kcne1 in lmpg micelles at ph 6.0 and2 40 degree c
Phyre2