| 1 |
|
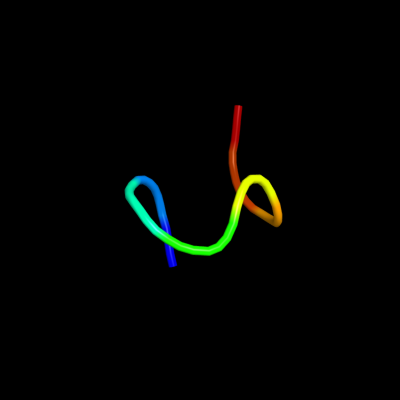
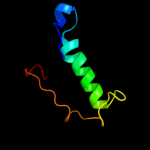
PDB 2iur chain D
Region: 19 - 31
Aligned: 13
Modelled: 13
Confidence: 9.9%
Identity: 54%
PDB header:oxidoreductase
Chain: D: PDB Molecule:aromatic amine dehydrogenase beta subunit;
PDBTitle: crystal structure of n-quinol form of aromatic amine2 dehydrogenase (aadh) from alcaligenes faecalis, form a3 cocrystal
Phyre2
| 2 |
|
PDB 2awi chain A domain 1
Region: 11 - 18
Aligned: 8
Modelled: 8
Confidence: 9.0%
Identity: 88%
Fold: lambda repressor-like DNA-binding domains
Superfamily: lambda repressor-like DNA-binding domains
Family: PrgX N-terminal domain-like
Phyre2
| 3 |
|
PDB 1prt chain C domain 2
Region: 54 - 65
Aligned: 12
Modelled: 12
Confidence: 7.4%
Identity: 50%
Fold: C-type lectin-like
Superfamily: C-type lectin-like
Family: Aerolysin/Pertussis toxin (APT) domain
Phyre2
| 4 |
|
PDB 1mda chain L
Region: 19 - 36
Aligned: 18
Modelled: 18
Confidence: 6.2%
Identity: 33%
Fold: Methylamine dehydrogenase, L chain
Superfamily: Methylamine dehydrogenase, L chain
Family: Methylamine dehydrogenase, L chain
Phyre2
| 5 |
|
PDB 3c75 chain L
Region: 19 - 36
Aligned: 18
Modelled: 18
Confidence: 5.5%
Identity: 22%
PDB header:oxidoreductase
Chain: L: PDB Molecule:methylamine dehydrogenase light chain;
PDBTitle: paracoccus versutus methylamine dehydrogenase in complex2 with amicyanin
Phyre2
| 6 |
|
PDB 1w36 chain B domain 1
Region: 27 - 84
Aligned: 50
Modelled: 58
Confidence: 5.5%
Identity: 26%
Fold: P-loop containing nucleoside triphosphate hydrolases
Superfamily: P-loop containing nucleoside triphosphate hydrolases
Family: Tandem AAA-ATPase domain
Phyre2
| 7 |
|
PDB 2bbk chain L
Region: 19 - 36
Aligned: 18
Modelled: 18
Confidence: 5.5%
Identity: 22%
Fold: Methylamine dehydrogenase, L chain
Superfamily: Methylamine dehydrogenase, L chain
Family: Methylamine dehydrogenase, L chain
Phyre2