| 1 |
|
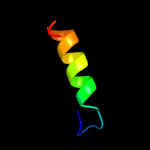
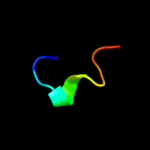
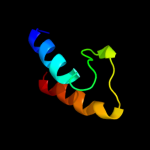
PDB 3c9p chain A
Region: 44 - 65
Aligned: 22
Modelled: 22
Confidence: 45.8%
Identity: 9%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein sp1917;
PDBTitle: crystal structure of uncharacterized protein sp1917
Phyre2
| 2 |
|
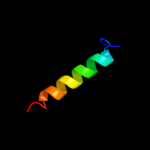
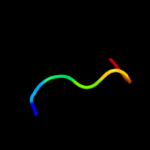
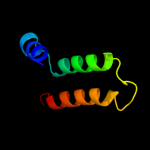
PDB 1a92 chain B
Region: 33 - 71
Aligned: 38
Modelled: 39
Confidence: 40.9%
Identity: 13%
PDB header:leucine zipper
Chain: B: PDB Molecule:delta antigen;
PDBTitle: oligomerization domain of hepatitis delta antigen
Phyre2
| 3 |
|
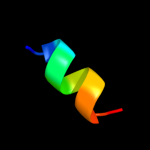
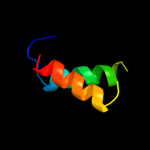
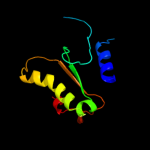
PDB 1k8i chain A domain 2
Region: 28 - 40
Aligned: 13
Modelled: 13
Confidence: 23.2%
Identity: 31%
Fold: MHC antigen-recognition domain
Superfamily: MHC antigen-recognition domain
Family: MHC antigen-recognition domain
Phyre2
| 4 |
|
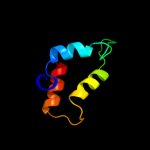
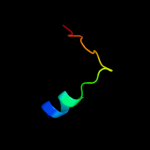
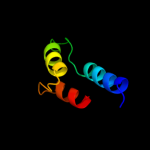
PDB 2bs2 chain A domain 1
Region: 107 - 122
Aligned: 16
Modelled: 16
Confidence: 13.4%
Identity: 38%
Fold: Spectrin repeat-like
Superfamily: Succinate dehydrogenase/fumarate reductase flavoprotein C-terminal domain
Family: Succinate dehydrogenase/fumarate reductase flavoprotein C-terminal domain
Phyre2
| 5 |
|
PDB 2fug chain 6 domain 1
Region: 45 - 67
Aligned: 23
Modelled: 23
Confidence: 10.5%
Identity: 9%
Fold: HydA/Nqo6-like
Superfamily: HydA/Nqo6-like
Family: Nq06-like
Phyre2
| 6 |
|
PDB 2jtv chain A
Region: 37 - 46
Aligned: 10
Modelled: 10
Confidence: 10.4%
Identity: 20%
PDB header:structural genomics
Chain: A: PDB Molecule:protein of unknown function;
PDBTitle: solution structure of protein rpa3401, northeast structural genomics2 consortium target rpt7, ontario center for structural proteomics3 target rp3384
Phyre2
| 7 |
|
PDB 3izc chain H
Region: 47 - 79
Aligned: 33
Modelled: 33
Confidence: 9.7%
Identity: 12%
PDB header:ribosome
Chain: H: PDB Molecule:60s ribosomal protein rpl8 (l7ae);
PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
Phyre2
| 8 |
|
PDB 2vvy chain C
Region: 4 - 63
Aligned: 50
Modelled: 60
Confidence: 9.4%
Identity: 16%
PDB header:viral protein
Chain: C: PDB Molecule:protein b15;
PDBTitle: structure of vaccinia virus protein b14
Phyre2
| 9 |
|
PDB 1hdm chain A domain 2
Region: 28 - 39
Aligned: 12
Modelled: 12
Confidence: 9.0%
Identity: 42%
Fold: MHC antigen-recognition domain
Superfamily: MHC antigen-recognition domain
Family: MHC antigen-recognition domain
Phyre2
| 10 |
|
PDB 1xkm chain C
Region: 114 - 120
Aligned: 7
Modelled: 7
Confidence: 8.8%
Identity: 43%
PDB header:antibiotic
Chain: C: PDB Molecule:distinctin chain a;
PDBTitle: nmr structure of antimicrobial peptide distinctin in water
Phyre2
| 11 |
|
PDB 1xkm chain A
Region: 114 - 120
Aligned: 7
Modelled: 7
Confidence: 8.7%
Identity: 43%
PDB header:antibiotic
Chain: A: PDB Molecule:distinctin chain a;
PDBTitle: nmr structure of antimicrobial peptide distinctin in water
Phyre2
| 12 |
|
PDB 2bpb chain B
Region: 27 - 64
Aligned: 38
Modelled: 38
Confidence: 7.8%
Identity: 11%
PDB header:oxidoreductase
Chain: B: PDB Molecule:sulfite\:cytochrome c oxidoreductase subunit b;
PDBTitle: sulfite dehydrogenase from starkeya novella
Phyre2
| 13 |
|
PDB 2gli chain A domain 5
Region: 74 - 83
Aligned: 10
Modelled: 10
Confidence: 7.2%
Identity: 40%
Fold: beta-beta-alpha zinc fingers
Superfamily: beta-beta-alpha zinc fingers
Family: Classic zinc finger, C2H2
Phyre2
| 14 |
|
PDB 2d35 chain A
Region: 34 - 42
Aligned: 9
Modelled: 9
Confidence: 6.6%
Identity: 56%
PDB header:cell cycle
Chain: A: PDB Molecule:cell division activator ceda;
PDBTitle: solution structure of cell division reactivation factor,2 ceda
Phyre2
| 15 |
|
PDB 2bn8 chain A
Region: 34 - 42
Aligned: 9
Modelled: 9
Confidence: 6.2%
Identity: 56%
PDB header:cell cycle protein
Chain: A: PDB Molecule:cell division activator ceda;
PDBTitle: solution structure and interactions of the e.coli cell2 division activator protein ceda
Phyre2
| 16 |
|
PDB 3e0z chain B
Region: 52 - 71
Aligned: 20
Modelled: 20
Confidence: 5.9%
Identity: 15%
PDB header:unknown function
Chain: B: PDB Molecule:protein of unknown function;
PDBTitle: crystal structure of a putative imidazole glycerol phosphate synthase2 homolog (eubrec_1070) from eubacterium rectale at 1.75 a resolution
Phyre2
| 17 |
|
PDB 1nsa chain A domain 2
Region: 49 - 89
Aligned: 41
Modelled: 41
Confidence: 5.6%
Identity: 12%
Fold: Ferredoxin-like
Superfamily: Protease propeptides/inhibitors
Family: Pancreatic carboxypeptidase, activation domain
Phyre2
| 18 |
|
PDB 3rys chain A
Region: 37 - 91
Aligned: 53
Modelled: 55
Confidence: 5.5%
Identity: 8%
PDB header:hydrolase
Chain: A: PDB Molecule:adenosine deaminase 1;
PDBTitle: the crystal structure of adenine deaminase (aaur1117) from2 arthrobacter aurescens
Phyre2
| 19 |
|
PDB 2y92 chain A
Region: 34 - 122
Aligned: 81
Modelled: 89
Confidence: 5.3%
Identity: 11%
PDB header:immune system
Chain: A: PDB Molecule:toll/interleukin-1 receptor domain-containing adapter
PDBTitle: crystal structure of mal adaptor protein
Phyre2
| 20 |
|
PDB 3ngv chain A
Region: 49 - 100
Aligned: 50
Modelled: 52
Confidence: 5.1%
Identity: 14%
PDB header:transport protein
Chain: A: PDB Molecule:d7 protein;
PDBTitle: crystal structure of anst-d7l1
Phyre2