| 1 |
|
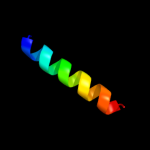
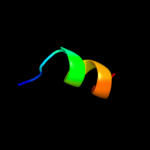
PDB 1jva chain A domain 2
Region: 16 - 24
Aligned: 9
Modelled: 9
Confidence: 35.4%
Identity: 67%
Fold: Homing endonuclease-like
Superfamily: Homing endonucleases
Family: Intein endonuclease
Phyre2
| 2 |
|
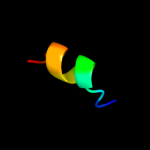
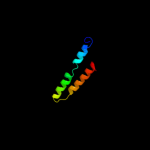
PDB 1q90 chain L
Region: 5 - 24
Aligned: 20
Modelled: 20
Confidence: 21.9%
Identity: 25%
Fold: Single transmembrane helix
Superfamily: PetL subunit of the cytochrome b6f complex
Family: PetL subunit of the cytochrome b6f complex
Phyre2
| 3 |
|
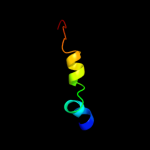
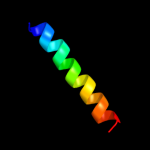
PDB 1q90 chain L
Region: 5 - 24
Aligned: 20
Modelled: 20
Confidence: 21.9%
Identity: 25%
PDB header:photosynthesis
Chain: L: PDB Molecule:cytochrome b6f complex subunit petl;
PDBTitle: structure of the cytochrome b6f (plastohydroquinone : plastocyanin2 oxidoreductase) from chlamydomonas reinhardtii
Phyre2
| 4 |
|
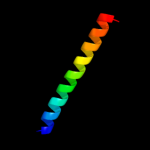
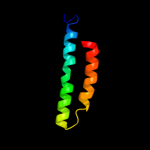
PDB 1b24 chain A
Region: 13 - 27
Aligned: 15
Modelled: 15
Confidence: 21.0%
Identity: 47%
PDB header:intron-encoded
Chain: A: PDB Molecule:protein (i-dmoi);
PDBTitle: i-dmoi, intron-encoded endonuclease
Phyre2
| 5 |
|
PDB 1b24 chain A domain 1
Region: 13 - 23
Aligned: 11
Modelled: 11
Confidence: 18.6%
Identity: 55%
Fold: Homing endonuclease-like
Superfamily: Homing endonucleases
Family: Group I mobile intron endonuclease
Phyre2
| 6 |
|
PDB 2i5n chain L domain 1
Region: 28 - 83
Aligned: 56
Modelled: 56
Confidence: 14.3%
Identity: 13%
Fold: Bacterial photosystem II reaction centre, L and M subunits
Superfamily: Bacterial photosystem II reaction centre, L and M subunits
Family: Bacterial photosystem II reaction centre, L and M subunits
Phyre2
| 7 |
|
PDB 3a0b chain X
Region: 86 - 96
Aligned: 11
Modelled: 11
Confidence: 11.1%
Identity: 9%
PDB header:electron transport
Chain: X: PDB Molecule:photosystem ii reaction center protein x;
PDBTitle: crystal structure of br-substituted photosystem ii complex
Phyre2
| 8 |
|
PDB 3a0b chain X
Region: 86 - 96
Aligned: 11
Modelled: 11
Confidence: 11.1%
Identity: 9%
PDB header:electron transport
Chain: X: PDB Molecule:photosystem ii reaction center protein x;
PDBTitle: crystal structure of br-substituted photosystem ii complex
Phyre2
| 9 |
|
PDB 3a0h chain X
Region: 86 - 96
Aligned: 11
Modelled: 11
Confidence: 11.1%
Identity: 9%
PDB header:electron transport
Chain: X: PDB Molecule:photosystem ii reaction center protein x;
PDBTitle: crystal structure of i-substituted photosystem ii complex
Phyre2
| 10 |
|
PDB 3a0h chain X
Region: 86 - 96
Aligned: 11
Modelled: 11
Confidence: 11.1%
Identity: 9%
PDB header:electron transport
Chain: X: PDB Molecule:photosystem ii reaction center protein x;
PDBTitle: crystal structure of i-substituted photosystem ii complex
Phyre2
| 11 |
|
PDB 2k21 chain A
Region: 36 - 63
Aligned: 26
Modelled: 28
Confidence: 10.5%
Identity: 31%
PDB header:membrane protein
Chain: A: PDB Molecule:potassium voltage-gated channel subfamily e
PDBTitle: nmr structure of human kcne1 in lmpg micelles at ph 6.0 and2 40 degree c
Phyre2
| 12 |
|
PDB 3rko chain F
Region: 39 - 59
Aligned: 21
Modelled: 21
Confidence: 9.8%
Identity: 19%
PDB header:oxidoreductase
Chain: F: PDB Molecule:nadh-quinone oxidoreductase subunit j;
PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
Phyre2
| 13 |
|
PDB 1m56 chain D
Region: 53 - 85
Aligned: 33
Modelled: 33
Confidence: 9.8%
Identity: 18%
Fold: Single transmembrane helix
Superfamily: Bacterial aa3 type cytochrome c oxidase subunit IV
Family: Bacterial aa3 type cytochrome c oxidase subunit IV
Phyre2
| 14 |
|
PDB 1s5l chain X
Region: 86 - 96
Aligned: 11
Modelled: 11
Confidence: 9.6%
Identity: 9%
PDB header:photosynthesis
Chain: X: PDB Molecule:photosystem ii psbx protein;
PDBTitle: architecture of the photosynthetic oxygen evolving center
Phyre2
| 15 |
|
PDB 1qle chain D
Region: 53 - 85
Aligned: 33
Modelled: 33
Confidence: 8.8%
Identity: 15%
Fold: Single transmembrane helix
Superfamily: Bacterial aa3 type cytochrome c oxidase subunit IV
Family: Bacterial aa3 type cytochrome c oxidase subunit IV
Phyre2
| 16 |
|
PDB 1eys chain M
Region: 28 - 83
Aligned: 56
Modelled: 56
Confidence: 8.5%
Identity: 21%
Fold: Bacterial photosystem II reaction centre, L and M subunits
Superfamily: Bacterial photosystem II reaction centre, L and M subunits
Family: Bacterial photosystem II reaction centre, L and M subunits
Phyre2
| 17 |
|
PDB 2j8c chain M domain 1
Region: 27 - 84
Aligned: 58
Modelled: 58
Confidence: 8.2%
Identity: 12%
Fold: Bacterial photosystem II reaction centre, L and M subunits
Superfamily: Bacterial photosystem II reaction centre, L and M subunits
Family: Bacterial photosystem II reaction centre, L and M subunits
Phyre2
| 18 |
|
PDB 2hac chain A
Region: 27 - 54
Aligned: 23
Modelled: 23
Confidence: 7.0%
Identity: 48%
PDB header:membrane protein
Chain: A: PDB Molecule:t-cell surface glycoprotein cd3 zeta chain;
PDBTitle: structure of zeta-zeta transmembrane dimer
Phyre2
| 19 |
|
PDB 2e0i chain D
Region: 23 - 35
Aligned: 13
Modelled: 13
Confidence: 7.0%
Identity: 23%
PDB header:lyase
Chain: D: PDB Molecule:432aa long hypothetical deoxyribodipyrimidine photolyase;
PDBTitle: crystal structure of archaeal photolyase from sulfolobus tokodaii with2 two fad molecules: implication of a novel light-harvesting cofactor
Phyre2
| 20 |
|
PDB 2j8c chain L domain 1
Region: 28 - 83
Aligned: 56
Modelled: 56
Confidence: 6.6%
Identity: 18%
Fold: Bacterial photosystem II reaction centre, L and M subunits
Superfamily: Bacterial photosystem II reaction centre, L and M subunits
Family: Bacterial photosystem II reaction centre, L and M subunits
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|