| 1 |
|
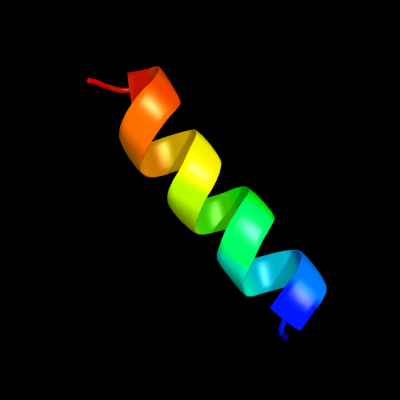
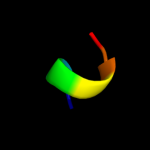
PDB 2jxf chain A
Region: 97 - 112
Aligned: 16
Modelled: 16
Confidence: 29.9%
Identity: 44%
PDB header:viral protein, membrane protein
Chain: A: PDB Molecule:genome polyprotein;
PDBTitle: the solution structure of hcv ns4b(40-69)
Phyre2
| 2 |
|
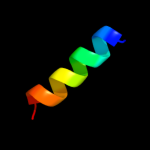
PDB 2jb0 chain B domain 1
Region: 83 - 92
Aligned: 10
Modelled: 10
Confidence: 16.5%
Identity: 60%
Fold: His-Me finger endonucleases
Superfamily: His-Me finger endonucleases
Family: HNH-motif
Phyre2
| 3 |
|
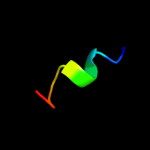
PDB 7cei chain B
Region: 83 - 92
Aligned: 10
Modelled: 10
Confidence: 15.5%
Identity: 60%
PDB header:immune system
Chain: B: PDB Molecule:protein (colicin e7 immunity protein);
PDBTitle: the endonuclease domain of colicin e7 in complex with its inhibitor2 im7 protein
Phyre2
| 4 |
|
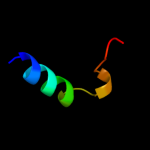
PDB 2gyk chain B domain 1
Region: 83 - 92
Aligned: 10
Modelled: 10
Confidence: 11.2%
Identity: 50%
Fold: His-Me finger endonucleases
Superfamily: His-Me finger endonucleases
Family: HNH-motif
Phyre2
| 5 |
|
PDB 1ukf chain A
Region: 93 - 101
Aligned: 9
Modelled: 9
Confidence: 7.8%
Identity: 56%
Fold: Cysteine proteinases
Superfamily: Cysteine proteinases
Family: Avirulence protein Avrpph3
Phyre2
| 6 |
|
PDB 1s4n chain A
Region: 67 - 89
Aligned: 23
Modelled: 23
Confidence: 7.7%
Identity: 35%
Fold: Nucleotide-diphospho-sugar transferases
Superfamily: Nucleotide-diphospho-sugar transferases
Family: Glycolipid 2-alpha-mannosyltransferase
Phyre2
| 7 |
|
PDB 2i68 chain B
Region: 99 - 104
Aligned: 6
Modelled: 6
Confidence: 6.1%
Identity: 67%
PDB header:transport protein
Chain: B: PDB Molecule:protein emre;
PDBTitle: cryo-em based theoretical model structure of transmembrane2 domain of the multidrug-resistance antiporter from e. coli3 emre
Phyre2
| 8 |
|
PDB 2ju0 chain B
Region: 57 - 68
Aligned: 12
Modelled: 12
Confidence: 6.1%
Identity: 42%
PDB header:metal binding protein/signaling protein
Chain: B: PDB Molecule:phosphatidylinositol 4-kinase pik1;
PDBTitle: structure of yeast frequenin bound to pdtins 4-kinase
Phyre2
| 9 |
|
PDB 2hac chain A
Region: 49 - 64
Aligned: 16
Modelled: 16
Confidence: 5.7%
Identity: 31%
PDB header:membrane protein
Chain: A: PDB Molecule:t-cell surface glycoprotein cd3 zeta chain;
PDBTitle: structure of zeta-zeta transmembrane dimer
Phyre2