| 1 |
|
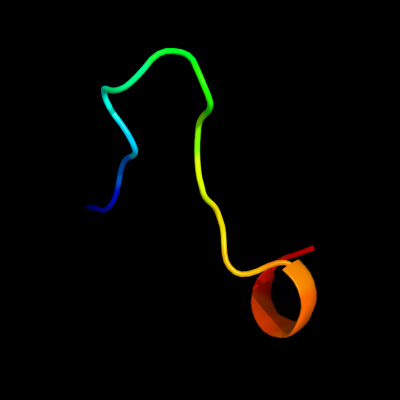
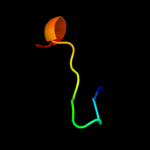
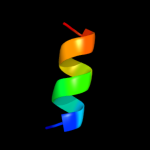
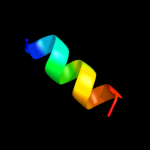
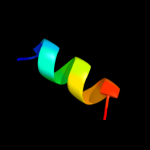
PDB 1v29 chain A
Region: 8 - 24
Aligned: 16
Modelled: 17
Confidence: 23.7%
Identity: 31%
Fold: Nitrile hydratase alpha chain
Superfamily: Nitrile hydratase alpha chain
Family: Nitrile hydratase alpha chain
Phyre2
| 2 |
|
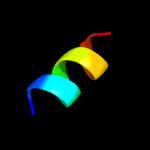
PDB 3mp7 chain B
Region: 56 - 77
Aligned: 21
Modelled: 22
Confidence: 23.5%
Identity: 38%
PDB header:protein transport
Chain: B: PDB Molecule:preprotein translocase subunit sece;
PDBTitle: lateral opening of a translocon upon entry of protein suggests the2 mechanism of insertion into membranes
Phyre2
| 3 |
|
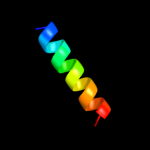
PDB 2f93 chain B domain 1
Region: 27 - 38
Aligned: 12
Modelled: 12
Confidence: 15.0%
Identity: 50%
Fold: Transmembrane helix hairpin
Superfamily: Htr2 transmembrane domain-like
Family: Htr2 transmembrane domain-like
Phyre2
| 4 |
|
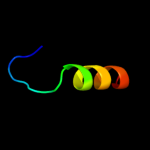
PDB 2f93 chain B
Region: 27 - 38
Aligned: 12
Modelled: 12
Confidence: 15.0%
Identity: 50%
PDB header:membrane protein
Chain: B: PDB Molecule:sensory rhodopsin ii transducer;
PDBTitle: k intermediate structure of sensory rhodopsin ii/transducer complex in2 combination with the ground state structure
Phyre2
| 5 |
|
PDB 2f95 chain B
Region: 27 - 38
Aligned: 12
Modelled: 12
Confidence: 13.8%
Identity: 50%
PDB header:membrane protein
Chain: B: PDB Molecule:sensory rhodopsin ii transducer;
PDBTitle: m intermediate structure of sensory rhodopsin ii/transducer complex in2 combination with the ground state structure
Phyre2
| 6 |
|
PDB 1h2s chain B
Region: 27 - 38
Aligned: 12
Modelled: 12
Confidence: 11.6%
Identity: 50%
Fold: Transmembrane helix hairpin
Superfamily: Htr2 transmembrane domain-like
Family: Htr2 transmembrane domain-like
Phyre2
| 7 |
|
PDB 1h2s chain B
Region: 27 - 38
Aligned: 12
Modelled: 12
Confidence: 11.6%
Identity: 50%
PDB header:membrane protein
Chain: B: PDB Molecule:sensory rhodopsin ii transducer;
PDBTitle: molecular basis of transmenbrane signalling by sensory2 rhodopsin ii-transducer complex
Phyre2
| 8 |
|
PDB 2ftx chain B domain 1
Region: 15 - 24
Aligned: 10
Modelled: 10
Confidence: 7.9%
Identity: 30%
Fold: Kinetochore globular domain-like
Superfamily: Kinetochore globular domain
Family: Spc24-like
Phyre2
| 9 |
|
PDB 2fv4 chain B
Region: 15 - 24
Aligned: 10
Modelled: 10
Confidence: 7.8%
Identity: 30%
PDB header:structural protein, protein binding
Chain: B: PDB Molecule:hypothetical 24.6 kda protein in ilv2-ade17
PDBTitle: nmr solution structure of the yeast kinetochore spc24/spc252 globular domain
Phyre2
| 10 |
|
PDB 2k1a chain A
Region: 25 - 43
Aligned: 19
Modelled: 19
Confidence: 5.8%
Identity: 21%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: bicelle-embedded integrin alpha(iib) transmembrane segment
Phyre2
| 11 |
|
PDB 1rzh chain H domain 2
Region: 25 - 45
Aligned: 19
Modelled: 21
Confidence: 5.3%
Identity: 47%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction centre subunit H, transmembrane region
Family: Photosystem II reaction centre subunit H, transmembrane region
Phyre2
| 12 |
|
PDB 2p41 chain A domain 1
Region: 14 - 33
Aligned: 20
Modelled: 20
Confidence: 5.3%
Identity: 25%
Fold: S-adenosyl-L-methionine-dependent methyltransferases
Superfamily: S-adenosyl-L-methionine-dependent methyltransferases
Family: mRNA cap methylase
Phyre2