| 1 |
|
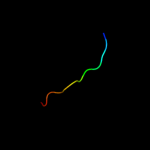
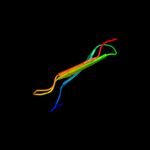
PDB 2h7a chain A domain 1
Region: 39 - 51
Aligned: 13
Modelled: 13
Confidence: 16.4%
Identity: 31%
Fold: YcgL-like
Superfamily: YcgL-like
Family: YcgL-like
Phyre2
| 2 |
|
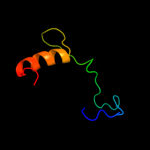
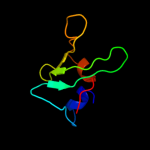
PDB 1sqr chain A
Region: 39 - 51
Aligned: 12
Modelled: 13
Confidence: 10.2%
Identity: 25%
Fold: Reductase/isomerase/elongation factor common domain
Superfamily: Translation proteins
Family: Ribosomal protein L35ae
Phyre2
| 3 |
|
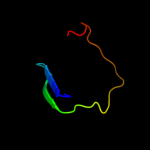
PDB 2l5q chain A
Region: 36 - 83
Aligned: 46
Modelled: 48
Confidence: 8.9%
Identity: 22%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of bvu_3817 from bacteroides vulgatus,2 northeast structural genomics consortium target bvr159
Phyre2
| 4 |
|
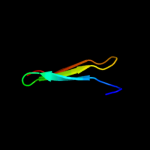
PDB 3g6n chain A
Region: 40 - 76
Aligned: 37
Modelled: 37
Confidence: 8.1%
Identity: 35%
PDB header:hydrolase
Chain: A: PDB Molecule:peptide deformylase;
PDBTitle: crystal structure of an efpdf complex with met-ala-ser
Phyre2
| 5 |
|
PDB 1msv chain B
Region: 39 - 64
Aligned: 26
Modelled: 26
Confidence: 6.8%
Identity: 31%
PDB header:lyase
Chain: B: PDB Molecule:s-adenosylmethionine decarboxylase proenzyme;
PDBTitle: the s68a s-adenosylmethionine decarboxylase proenzyme2 processing mutant.
Phyre2
| 6 |
|
PDB 1jl0 chain A
Region: 39 - 64
Aligned: 26
Modelled: 26
Confidence: 6.4%
Identity: 35%
Fold: S-adenosylmethionine decarboxylase
Superfamily: S-adenosylmethionine decarboxylase
Family: S-adenosylmethionine decarboxylase
Phyre2
| 7 |
|
PDB 2k2o chain A
Region: 33 - 51
Aligned: 19
Modelled: 19
Confidence: 6.2%
Identity: 32%
PDB header:membrane protein
Chain: A: PDB Molecule:myoferlin;
PDBTitle: solution structure of the inner dysf domain of human2 myoferlin
Phyre2
| 8 |
|
PDB 2bjq chain A domain 1
Region: 13 - 76
Aligned: 64
Modelled: 64
Confidence: 6.1%
Identity: 16%
Fold: MFPT repeat-like
Superfamily: MFPT repeat-like
Family: MFPT repeat
Phyre2
| 9 |
|
PDB 2zgw chain A domain 1
Region: 7 - 15
Aligned: 9
Modelled: 9
Confidence: 5.4%
Identity: 56%
Fold: SH3-like barrel
Superfamily: C-terminal domain of transcriptional repressors
Family: Biotin repressor (BirA)
Phyre2