| 1 |
|
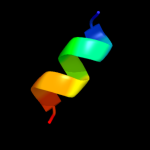
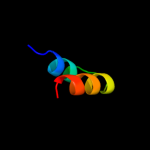
PDB 1xhm chain B domain 1
Region: 75 - 85
Aligned: 11
Modelled: 11
Confidence: 30.5%
Identity: 36%
Fold: Non-globular all-alpha subunits of globular proteins
Superfamily: Transducin (heterotrimeric G protein), gamma chain
Family: Transducin (heterotrimeric G protein), gamma chain
Phyre2
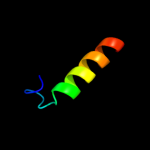
| 2 |
|
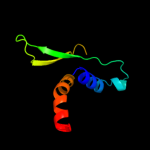
PDB 1xhm chain B
Region: 75 - 85
Aligned: 11
Modelled: 11
Confidence: 30.4%
Identity: 36%
PDB header:signaling protein
Chain: B: PDB Molecule:guanine nucleotide-binding protein g(i)/g(s)
PDBTitle: the crystal structure of a biologically active peptide2 (sigk) bound to a g protein beta:gamma heterodimer
Phyre2
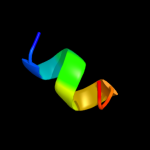
| 3 |
|
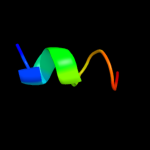
PDB 2kel chain B
Region: 81 - 103
Aligned: 23
Modelled: 23
Confidence: 26.5%
Identity: 35%
PDB header:transcription repressor
Chain: B: PDB Molecule:uncharacterized protein 56b;
PDBTitle: structure of the transcription regulator svtr from the2 hyperthermophilic archaeal virus sirv1
Phyre2
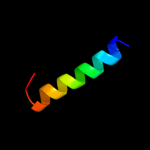
| 4 |
|
PDB 1elk chain A
Region: 78 - 85
Aligned: 8
Modelled: 8
Confidence: 11.0%
Identity: 38%
Fold: alpha-alpha superhelix
Superfamily: ENTH/VHS domain
Family: VHS domain
Phyre2
| 5 |
|
PDB 1xb4 chain A domain 1
Region: 78 - 87
Aligned: 10
Modelled: 10
Confidence: 10.6%
Identity: 50%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: Vacuolar sorting protein domain
Phyre2
| 6 |
|
PDB 1g8m chain A domain 2
Region: 85 - 107
Aligned: 23
Modelled: 23
Confidence: 8.9%
Identity: 17%
Fold: Cytidine deaminase-like
Superfamily: Cytidine deaminase-like
Family: AICAR transformylase domain of bifunctional purine biosynthesis enzyme ATIC
Phyre2
| 7 |
|
PDB 2ahm chain A domain 1
Region: 80 - 104
Aligned: 25
Modelled: 25
Confidence: 8.4%
Identity: 12%
Fold: immunoglobulin/albumin-binding domain-like
Superfamily: Coronavirus NSP7-like
Family: Coronavirus NSP7-like
Phyre2
| 8 |
|
PDB 2zme chain D
Region: 78 - 86
Aligned: 9
Modelled: 9
Confidence: 7.7%
Identity: 33%
PDB header:protein transport
Chain: D: PDB Molecule:vacuolar protein-sorting-associated protein 25;
PDBTitle: integrated structural and functional model of the human escrt-ii2 complex
Phyre2
| 9 |
|
PDB 1ysy chain A domain 1
Region: 80 - 104
Aligned: 25
Modelled: 25
Confidence: 7.4%
Identity: 12%
Fold: immunoglobulin/albumin-binding domain-like
Superfamily: Coronavirus NSP7-like
Family: Coronavirus NSP7-like
Phyre2
| 10 |
|
PDB 1ldm chain A domain 2
Region: 92 - 159
Aligned: 60
Modelled: 68
Confidence: 7.3%
Identity: 22%
Fold: LDH C-terminal domain-like
Superfamily: LDH C-terminal domain-like
Family: Lactate & malate dehydrogenases, C-terminal domain
Phyre2
| 11 |
|
PDB 3rru chain A
Region: 78 - 85
Aligned: 8
Modelled: 8
Confidence: 6.7%
Identity: 13%
PDB header:signaling protein
Chain: A: PDB Molecule:tom1l1 protein;
PDBTitle: x-ray crystal structure of the vhs domain of human tom1-like protein,2 northeast structural genomics consortium target hr3050e
Phyre2
| 12 |
|
PDB 1pkx chain A domain 2
Region: 85 - 107
Aligned: 23
Modelled: 23
Confidence: 6.6%
Identity: 22%
Fold: Cytidine deaminase-like
Superfamily: Cytidine deaminase-like
Family: AICAR transformylase domain of bifunctional purine biosynthesis enzyme ATIC
Phyre2
| 13 |
|
PDB 3cuq chain D
Region: 78 - 86
Aligned: 9
Modelled: 9
Confidence: 6.4%
Identity: 33%
PDB header:protein transport
Chain: D: PDB Molecule:vacuolar protein-sorting-associated protein 25;
PDBTitle: integrated structural and functional model of the human escrt-ii2 complex
Phyre2
| 14 |
|
PDB 1mwp chain A
Region: 77 - 84
Aligned: 8
Modelled: 8
Confidence: 6.4%
Identity: 75%
Fold: SRCR-like
Superfamily: A heparin-binding domain
Family: A heparin-binding domain
Phyre2
| 15 |
|
PDB 3c2q chain A
Region: 77 - 87
Aligned: 11
Modelled: 11
Confidence: 5.9%
Identity: 36%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized conserved protein;
PDBTitle: crystal structure of conserved putative lor/sdh protein2 from methanococcus maripaludis s2
Phyre2
| 16 |
|
PDB 1omw chain G
Region: 75 - 85
Aligned: 11
Modelled: 11
Confidence: 5.9%
Identity: 36%
Fold: Non-globular all-alpha subunits of globular proteins
Superfamily: Transducin (heterotrimeric G protein), gamma chain
Family: Transducin (heterotrimeric G protein), gamma chain
Phyre2
| 17 |
|
PDB 1mhq chain A
Region: 78 - 85
Aligned: 8
Modelled: 8
Confidence: 5.7%
Identity: 25%
Fold: alpha-alpha superhelix
Superfamily: ENTH/VHS domain
Family: VHS domain
Phyre2
| 18 |
|
PDB 1ceu chain A
Region: 75 - 85
Aligned: 11
Modelled: 11
Confidence: 5.5%
Identity: 9%
PDB header:viral protein
Chain: A: PDB Molecule:protein (hiv-1 regulatory protein n-terminal
PDBTitle: nmr structure of the (1-51) n-terminal domain of the hiv-12 regulatory protein
Phyre2