| 1 |
|
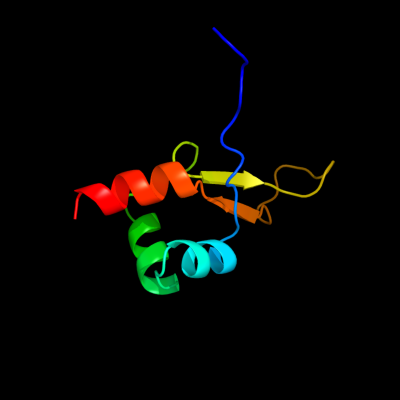
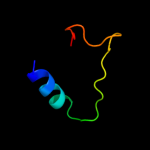
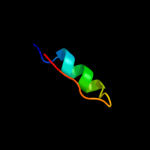
PDB 2kvv chain A
Region: 1 - 71
Aligned: 70
Modelled: 71
Confidence: 100.0%
Identity: 73%
PDB header:hydrolase
Chain: A: PDB Molecule:putative excisionase;
PDBTitle: solution nmr of putative excisionase from klebsiella pneumoniae,2 northeast structural genomics consortium target target kpr49
Phyre2
| 2 |
|
PDB 2fok chain A domain 4
Region: 36 - 59
Aligned: 21
Modelled: 21
Confidence: 14.0%
Identity: 38%
Fold: Restriction endonuclease-like
Superfamily: Restriction endonuclease-like
Family: Restriction endonuclease FokI, C-terminal (catalytic) domain
Phyre2
| 3 |
|
PDB 3pc3 chain A
Region: 5 - 23
Aligned: 19
Modelled: 19
Confidence: 13.0%
Identity: 11%
PDB header:lyase
Chain: A: PDB Molecule:cg1753, isoform a;
PDBTitle: full length structure of cystathionine beta-synthase from drosophila2 in complex with aminoacrylate
Phyre2
| 4 |
|
PDB 1ffk chain F
Region: 22 - 52
Aligned: 31
Modelled: 31
Confidence: 9.3%
Identity: 29%
Fold: alpha/beta-Hammerhead
Superfamily: Ribosomal protein L16p/L10e
Family: Ribosomal protein L10e
Phyre2
| 5 |
|
PDB 1efe chain A
Region: 23 - 56
Aligned: 34
Modelled: 34
Confidence: 9.1%
Identity: 32%
Fold: Insulin-like
Superfamily: Insulin-like
Family: Insulin-like
Phyre2
| 6 |
|
PDB 2ih2 chain A domain 2
Region: 23 - 70
Aligned: 47
Modelled: 48
Confidence: 8.6%
Identity: 21%
Fold: DNA methylase specificity domain
Superfamily: DNA methylase specificity domain
Family: TaqI C-terminal domain-like
Phyre2
| 7 |
|
PDB 1hp9 chain A
Region: 29 - 38
Aligned: 10
Modelled: 10
Confidence: 8.5%
Identity: 60%
PDB header:toxin
Chain: A: PDB Molecule:kappa-hefutoxin 1;
PDBTitle: kappa-hefutoxins: a novel class of potassium channel toxins2 from scorpion venom
Phyre2
| 8 |
|
PDB 2o2a chain A domain 1
Region: 27 - 46
Aligned: 20
Modelled: 19
Confidence: 6.6%
Identity: 20%
Fold: SecB-like
Superfamily: SecB-like
Family: SP1558-like
Phyre2
| 9 |
|
PDB 1liu chain A domain 3
Region: 19 - 39
Aligned: 21
Modelled: 21
Confidence: 6.2%
Identity: 29%
Fold: Pyruvate kinase C-terminal domain-like
Superfamily: PK C-terminal domain-like
Family: Pyruvate kinase, C-terminal domain
Phyre2
| 10 |
|
PDB 2lk2 chain A
Region: 16 - 55
Aligned: 40
Modelled: 40
Confidence: 5.8%
Identity: 23%
PDB header:transcription
Chain: A: PDB Molecule:homeobox protein tgif1;
PDBTitle: solution nmr structure of homeobox domain (171-248) of human homeobox2 protein tgif1, northeast structural genomics consortium target3 hr4411b
Phyre2
| 11 |
|
PDB 1a3x chain A domain 3
Region: 19 - 39
Aligned: 21
Modelled: 21
Confidence: 5.7%
Identity: 29%
Fold: Pyruvate kinase C-terminal domain-like
Superfamily: PK C-terminal domain-like
Family: Pyruvate kinase, C-terminal domain
Phyre2
| 12 |
|
PDB 2hng chain A domain 1
Region: 31 - 46
Aligned: 16
Modelled: 16
Confidence: 5.6%
Identity: 25%
Fold: SecB-like
Superfamily: SecB-like
Family: SP1558-like
Phyre2