| 1 |
|
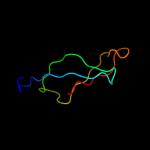
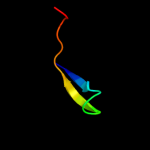
PDB 2zjr chain 1 domain 1
Region: 3 - 53
Aligned: 51
Modelled: 51
Confidence: 99.9%
Identity: 57%
Fold: Rubredoxin-like
Superfamily: Zn-binding ribosomal proteins
Family: Ribosomal protein L33p
Phyre2
| 2 |
|
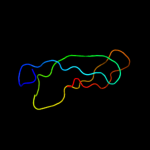
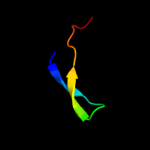
PDB 2gyc chain 1 domain 1
Region: 2 - 53
Aligned: 52
Modelled: 52
Confidence: 99.9%
Identity: 100%
Fold: Rubredoxin-like
Superfamily: Zn-binding ribosomal proteins
Family: Ribosomal protein L33p
Phyre2
| 3 |
|
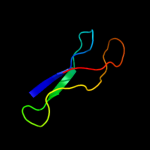
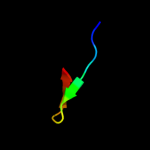
PDB 2b9n chain 6
Region: 3 - 53
Aligned: 51
Modelled: 51
Confidence: 99.9%
Identity: 57%
PDB header:ribosome
Chain: 6: PDB Molecule:50s ribosomal protein l33;
PDBTitle: 50s ribosomal subunit from a crystal structure of release factor rf2,2 trnas and mrna bound to the ribosome. this file contains the 50s3 subunit from a crystal structure of release factor rf1, trnas and4 mrna bound to the ribosome and is described in remark 400.
Phyre2
| 4 |
|
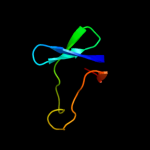
PDB 2ftc chain P
Region: 2 - 53
Aligned: 50
Modelled: 52
Confidence: 99.8%
Identity: 44%
PDB header:ribosome
Chain: P: PDB Molecule:mitochondrial ribosomal protein l33 isoform a;
PDBTitle: structural model for the large subunit of the mammalian mitochondrial2 ribosome
Phyre2
| 5 |
|
PDB 3bbo chain 3
Region: 1 - 53
Aligned: 53
Modelled: 53
Confidence: 99.5%
Identity: 42%
PDB header:ribosome
Chain: 3: PDB Molecule:ribosomal protein l33;
PDBTitle: homology model for the spinach chloroplast 50s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome
Phyre2
| 6 |
|
PDB 3d5d chain 6
Region: 9 - 51
Aligned: 43
Modelled: 43
Confidence: 98.9%
Identity: 40%
PDB header:ribosome
Chain: 6: PDB Molecule:50s ribosomal protein l33;
PDBTitle: structural basis for translation termination on the 70s ribosome. this2 file contains the 50s subunit of the second 70s ribosome. the entire3 crystal structure contains two 70s ribosomes as described in remark4 400.
Phyre2
| 7 |
|
PDB 3f1z chain F
Region: 3 - 33
Aligned: 31
Modelled: 31
Confidence: 23.5%
Identity: 29%
PDB header:dna binding protein
Chain: F: PDB Molecule:putative nucleic acid-binding lipoprotein;
PDBTitle: crystal structure of putative nucleic acid-binding lipoprotein2 (yp_001337197.1) from klebsiella pneumoniae subsp. pneumoniae mgh3 78578 at 2.46 a resolution
Phyre2
| 8 |
|
PDB 1a9x chain A domain 1
Region: 4 - 24
Aligned: 21
Modelled: 21
Confidence: 19.4%
Identity: 24%
Fold: Carbamoyl phosphate synthetase, large subunit connection domain
Superfamily: Carbamoyl phosphate synthetase, large subunit connection domain
Family: Carbamoyl phosphate synthetase, large subunit connection domain
Phyre2
| 9 |
|
PDB 1yfb chain A domain 1
Region: 32 - 41
Aligned: 10
Modelled: 10
Confidence: 13.8%
Identity: 60%
Fold: Double-split beta-barrel
Superfamily: AbrB/MazE/MraZ-like
Family: AbrB N-terminal domain-like
Phyre2
| 10 |
|
PDB 2fy9 chain A domain 1
Region: 32 - 41
Aligned: 10
Modelled: 10
Confidence: 9.4%
Identity: 50%
Fold: Double-split beta-barrel
Superfamily: AbrB/MazE/MraZ-like
Family: AbrB N-terminal domain-like
Phyre2
| 11 |
|
PDB 1vqo chain 3 domain 1
Region: 35 - 55
Aligned: 21
Modelled: 21
Confidence: 9.3%
Identity: 14%
Fold: Rubredoxin-like
Superfamily: Zn-binding ribosomal proteins
Family: Ribosomal protein L44e
Phyre2
| 12 |
|
PDB 1u0l chain A domain 1
Region: 10 - 49
Aligned: 40
Modelled: 40
Confidence: 8.9%
Identity: 20%
Fold: OB-fold
Superfamily: Nucleic acid-binding proteins
Family: Cold shock DNA-binding domain-like
Phyre2
| 13 |
|
PDB 2zkr chain 4
Region: 35 - 55
Aligned: 21
Modelled: 21
Confidence: 8.2%
Identity: 19%
PDB header:ribosomal protein/rna
Chain: 4: PDB Molecule:60s ribosomal protein l44e;
PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
Phyre2
| 14 |
|
PDB 2ro5 chain B
Region: 33 - 42
Aligned: 10
Modelled: 10
Confidence: 7.5%
Identity: 40%
PDB header:transcription
Chain: B: PDB Molecule:stage v sporulation protein t;
PDBTitle: rdc-refined solution structure of the n-terminal dna2 recognition domain of the bacillus subtilis transition-3 state regulator spovt
Phyre2
| 15 |
|
PDB 2w9j chain B
Region: 14 - 27
Aligned: 14
Modelled: 14
Confidence: 7.4%
Identity: 50%
PDB header:signaling protein
Chain: B: PDB Molecule:signal recognition particle subunit srp14;
PDBTitle: the crystal structure of srp14 from the schizosaccharomyces2 pombe signal recognition particle
Phyre2
| 16 |
|
PDB 4a19 chain C
Region: 35 - 55
Aligned: 21
Modelled: 21
Confidence: 7.3%
Identity: 19%
PDB header:ribosome
Chain: C: PDB Molecule:60s ribosomal protein l36a;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 26s rrna and3 proteins of molecule 2.
Phyre2
| 17 |
|
PDB 1fs1 chain B domain 2
Region: 7 - 19
Aligned: 13
Modelled: 13
Confidence: 6.5%
Identity: 46%
Fold: POZ domain
Superfamily: POZ domain
Family: BTB/POZ domain
Phyre2