| 1 |
|
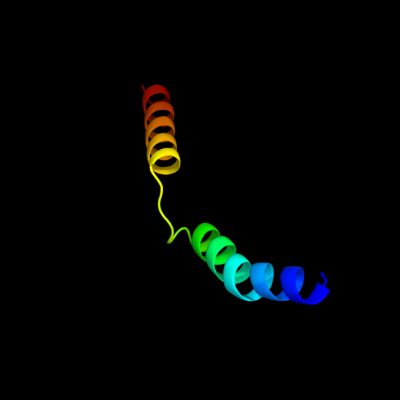
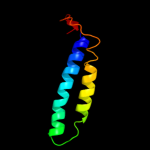
PDB 2oar chain A
Region: 85 - 130
Aligned: 46
Modelled: 46
Confidence: 28.8%
Identity: 13%
PDB header:membrane protein
Chain: A: PDB Molecule:large-conductance mechanosensitive channel;
PDBTitle: mechanosensitive channel of large conductance (mscl)
Phyre2
| 2 |
|
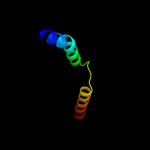
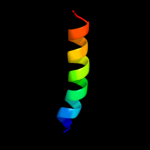
PDB 2z9f chain C
Region: 98 - 127
Aligned: 30
Modelled: 30
Confidence: 24.8%
Identity: 10%
PDB header:biosynthetic protein
Chain: C: PDB Molecule:cellulose synthase operon protein d;
PDBTitle: crystal structure of axcesd protein from acetobacter xylinum
Phyre2
| 3 |
|
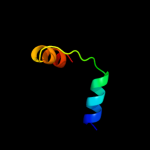
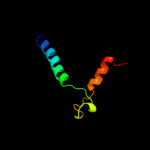
PDB 2knc chain A
Region: 80 - 113
Aligned: 34
Modelled: 34
Confidence: 8.3%
Identity: 15%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 4 |
|
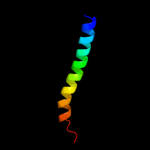
PDB 3pjz chain A
Region: 51 - 115
Aligned: 65
Modelled: 65
Confidence: 7.2%
Identity: 14%
PDB header:transport protein
Chain: A: PDB Molecule:potassium uptake protein trkh;
PDBTitle: crystal structure of the potassium transporter trkh from vibrio2 parahaemolyticus
Phyre2
| 5 |
|
PDB 2klu chain A
Region: 50 - 71
Aligned: 22
Modelled: 22
Confidence: 6.9%
Identity: 23%
PDB header:immune system, membrane protein
Chain: A: PDB Molecule:t-cell surface glycoprotein cd4;
PDBTitle: nmr structure of the transmembrane and cytoplasmic domains2 of human cd4
Phyre2
| 6 |
|
PDB 2jo1 chain A
Region: 77 - 132
Aligned: 56
Modelled: 56
Confidence: 6.8%
Identity: 9%
PDB header:hydrolase regulator
Chain: A: PDB Molecule:phospholemman;
PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
Phyre2
| 7 |
|
PDB 2bbj chain B
Region: 15 - 106
Aligned: 83
Modelled: 92
Confidence: 6.7%
Identity: 7%
PDB header:metal transport/membrane protein
Chain: B: PDB Molecule:divalent cation transport-related protein;
PDBTitle: crystal structure of the cora mg2+ transporter
Phyre2