| 1 |
|
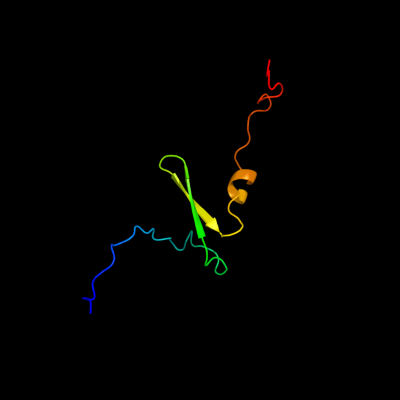
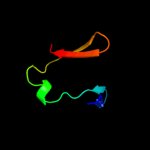
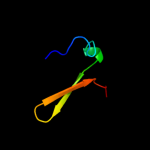
PDB 3tnd chain F
Region: 2 - 67
Aligned: 66
Modelled: 66
Confidence: 100.0%
Identity: 97%
PDB header:translation, toxin
Chain: F: PDB Molecule:antitoxin vapb;
PDBTitle: crystal structure of shigella flexneri vapbc toxin-antitoxin complex
Phyre2
| 2 |
|
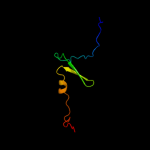
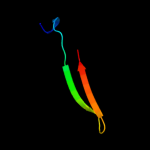
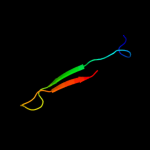
PDB 3zvk chain G
Region: 2 - 75
Aligned: 72
Modelled: 74
Confidence: 100.0%
Identity: 32%
PDB header:antitoxin/toxin/dna
Chain: G: PDB Molecule:antitoxin of toxin-antitoxin system vapb;
PDBTitle: crystal structure of vapbc2 from rickettsia felis bound to2 a dna fragment from their promoter
Phyre2
| 3 |
|
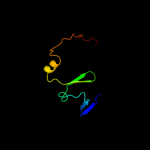
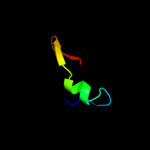
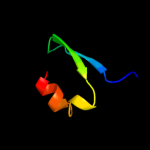
PDB 1ub4 chain C
Region: 1 - 55
Aligned: 53
Modelled: 55
Confidence: 98.9%
Identity: 28%
Fold: Double-split beta-barrel
Superfamily: AbrB/MazE/MraZ-like
Family: Kis/PemI addiction antidote
Phyre2
| 4 |
|
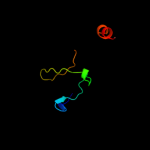
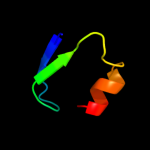
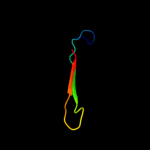
PDB 1mvf chain D
Region: 3 - 43
Aligned: 41
Modelled: 41
Confidence: 97.6%
Identity: 29%
Fold: Double-split beta-barrel
Superfamily: AbrB/MazE/MraZ-like
Family: Kis/PemI addiction antidote
Phyre2
| 5 |
|
PDB 1una chain A
Region: 8 - 34
Aligned: 27
Modelled: 27
Confidence: 15.1%
Identity: 30%
Fold: RNA bacteriophage capsid protein
Superfamily: RNA bacteriophage capsid protein
Family: RNA bacteriophage capsid protein
Phyre2
| 6 |
|
PDB 2fy9 chain A domain 1
Region: 14 - 42
Aligned: 27
Modelled: 29
Confidence: 14.0%
Identity: 19%
Fold: Double-split beta-barrel
Superfamily: AbrB/MazE/MraZ-like
Family: AbrB N-terminal domain-like
Phyre2
| 7 |
|
PDB 1uf0 chain A
Region: 26 - 54
Aligned: 29
Modelled: 29
Confidence: 10.5%
Identity: 21%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: Doublecortin (DC)
Family: Doublecortin (DC)
Phyre2
| 8 |
|
PDB 1yfb chain A domain 1
Region: 14 - 46
Aligned: 31
Modelled: 33
Confidence: 9.6%
Identity: 13%
Fold: Double-split beta-barrel
Superfamily: AbrB/MazE/MraZ-like
Family: AbrB N-terminal domain-like
Phyre2
| 9 |
|
PDB 3ila chain G
Region: 17 - 45
Aligned: 23
Modelled: 29
Confidence: 8.3%
Identity: 26%
PDB header:signaling protein
Chain: G: PDB Molecule:ryanodine receptor 1;
PDBTitle: crystal structure of rabbit ryanodine receptor 1 n-terminal domain (9-2 205)
Phyre2
| 10 |
|
PDB 2qc8 chain J
Region: 8 - 17
Aligned: 10
Modelled: 10
Confidence: 8.2%
Identity: 50%
PDB header:ligase
Chain: J: PDB Molecule:glutamine synthetase;
PDBTitle: crystal structure of human glutamine synthetase in complex with adp2 and methionine sulfoximine phosphate
Phyre2
| 11 |
|
PDB 2d3a chain J
Region: 8 - 20
Aligned: 13
Modelled: 13
Confidence: 6.7%
Identity: 31%
PDB header:ligase
Chain: J: PDB Molecule:glutamine synthetase;
PDBTitle: crystal structure of the maize glutamine synthetase2 complexed with adp and methionine sulfoximine phosphate
Phyre2
| 12 |
|
PDB 1fr5 chain A
Region: 8 - 34
Aligned: 27
Modelled: 27
Confidence: 6.7%
Identity: 26%
Fold: RNA bacteriophage capsid protein
Superfamily: RNA bacteriophage capsid protein
Family: RNA bacteriophage capsid protein
Phyre2
| 13 |
|
PDB 2hjq chain A domain 2
Region: 3 - 14
Aligned: 12
Modelled: 12
Confidence: 6.5%
Identity: 33%
Fold: GINS/PriA/YqbF domain
Superfamily: PriA/YqbF domain
Family: YqbF N-terminal domain-like
Phyre2
| 14 |
|
PDB 1mjd chain A
Region: 23 - 54
Aligned: 32
Modelled: 32
Confidence: 6.3%
Identity: 16%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: Doublecortin (DC)
Family: Doublecortin (DC)
Phyre2
| 15 |
|
PDB 2bu1 chain A domain 1
Region: 8 - 34
Aligned: 27
Modelled: 27
Confidence: 6.3%
Identity: 30%
Fold: RNA bacteriophage capsid protein
Superfamily: RNA bacteriophage capsid protein
Family: RNA bacteriophage capsid protein
Phyre2
| 16 |
|
PDB 2w1t chain B
Region: 14 - 46
Aligned: 31
Modelled: 33
Confidence: 6.2%
Identity: 16%
PDB header:transcription
Chain: B: PDB Molecule:stage v sporulation protein t;
PDBTitle: crystal structure of b. subtilis spovt
Phyre2
| 17 |
|
PDB 1mg4 chain A
Region: 27 - 54
Aligned: 28
Modelled: 28
Confidence: 6.0%
Identity: 21%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: Doublecortin (DC)
Family: Doublecortin (DC)
Phyre2
| 18 |
|
PDB 2a6q chain A domain 1
Region: 25 - 54
Aligned: 30
Modelled: 30
Confidence: 5.7%
Identity: 3%
Fold: YefM-like
Superfamily: YefM-like
Family: YefM-like
Phyre2
| 19 |
|
PDB 3bmv chain A domain 3
Region: 29 - 46
Aligned: 18
Modelled: 18
Confidence: 5.6%
Identity: 6%
Fold: Glycosyl hydrolase domain
Superfamily: Glycosyl hydrolase domain
Family: alpha-Amylases, C-terminal beta-sheet domain
Phyre2
| 20 |
|
PDB 1v5r chain A domain 1
Region: 20 - 54
Aligned: 33
Modelled: 35
Confidence: 5.2%
Identity: 9%
Fold: N domain of copper amine oxidase-like
Superfamily: GAS2 domain-like
Family: GAS2 domain
Phyre2
| 21 |
|
| 22 |
|