| 1 |
|
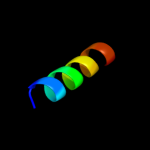
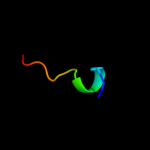
PDB 1bts chain A
Region: 52 - 67
Aligned: 16
Modelled: 16
Confidence: 32.3%
Identity: 50%
PDB header:transmembrane protein
Chain: A: PDB Molecule:band 3 anion transport protein;
PDBTitle: the solution structures of the first and second2 transmembrane-spanning segments of band 3
Phyre2
| 2 |
|
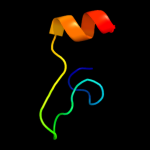
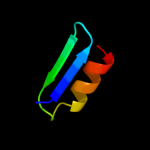
PDB 1btt chain A
Region: 52 - 67
Aligned: 16
Modelled: 16
Confidence: 32.3%
Identity: 50%
PDB header:transmembrane protein
Chain: A: PDB Molecule:band 3 anion transport protein;
PDBTitle: the solution structures of the first and second2 transmembrane-spanning segments of band 3
Phyre2
| 3 |
|
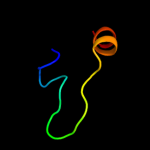
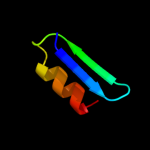
PDB 3etb chain J
Region: 5 - 13
Aligned: 9
Modelled: 9
Confidence: 25.5%
Identity: 67%
PDB header:immune system/toxin
Chain: J: PDB Molecule:anthrax protective antigen;
PDBTitle: crystal structure of the engineered neutralizing antibody2 m18 complexed with anthrax protective antigen domain 4
Phyre2
| 4 |
|
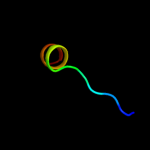
PDB 2zme chain D
Region: 42 - 66
Aligned: 25
Modelled: 25
Confidence: 9.7%
Identity: 32%
PDB header:protein transport
Chain: D: PDB Molecule:vacuolar protein-sorting-associated protein 25;
PDBTitle: integrated structural and functional model of the human escrt-ii2 complex
Phyre2
| 5 |
|
PDB 3qte chain A
Region: 62 - 69
Aligned: 8
Modelled: 8
Confidence: 7.8%
Identity: 63%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:defensin-6;
PDBTitle: crystal structure of human alpha-defensin 6 (h27w mutant)
Phyre2
| 6 |
|
PDB 3cuq chain D
Region: 42 - 66
Aligned: 25
Modelled: 25
Confidence: 7.0%
Identity: 32%
PDB header:protein transport
Chain: D: PDB Molecule:vacuolar protein-sorting-associated protein 25;
PDBTitle: integrated structural and functional model of the human escrt-ii2 complex
Phyre2
| 7 |
|
PDB 2fhd chain A
Region: 15 - 27
Aligned: 13
Modelled: 13
Confidence: 6.4%
Identity: 38%
PDB header:cell cycle
Chain: A: PDB Molecule:dna repair protein rhp9/crb2;
PDBTitle: crystal structure of crb2 tandem tudor domains
Phyre2
| 8 |
|
PDB 1xb4 chain A domain 1
Region: 53 - 66
Aligned: 14
Modelled: 14
Confidence: 6.2%
Identity: 14%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: Vacuolar sorting protein domain
Phyre2
| 9 |
|
PDB 2a06 chain V
Region: 1 - 12
Aligned: 12
Modelled: 12
Confidence: 6.0%
Identity: 58%
PDB header:oxidoreductase
Chain: V: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur subunit,
PDBTitle: bovine cytochrome bc1 complex with stigmatellin bound
Phyre2
| 10 |
|
PDB 1rpu chain A
Region: 18 - 45
Aligned: 28
Modelled: 28
Confidence: 5.7%
Identity: 43%
Fold: Tombusvirus P19 core protein, VP19
Superfamily: Tombusvirus P19 core protein, VP19
Family: Tombusvirus P19 core protein, VP19
Phyre2
| 11 |
|
PDB 1rpu chain A
Region: 18 - 45
Aligned: 28
Modelled: 28
Confidence: 5.7%
Identity: 43%
PDB header:rna binding protein/rna
Chain: A: PDB Molecule:19 kda protein;
PDBTitle: crystal structure of cirv p19 bound to sirna
Phyre2
| 12 |
|
PDB 1ppj chain I
Region: 1 - 12
Aligned: 12
Modelled: 12
Confidence: 5.5%
Identity: 58%
Fold: Non-globular alpha+beta subunits of globular proteins
Superfamily: Non-globular alpha+beta subunits of globular proteins
Family: Ubiquinol-cytochrome c reductase 8 kDa protein
Phyre2
| 13 |
|
PDB 1zej chain A
Region: 7 - 21
Aligned: 15
Modelled: 15
Confidence: 5.4%
Identity: 27%
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-hydroxyacyl-coa dehydrogenase;
PDBTitle: crystal structure of the 3-hydroxyacyl-coa dehydrogenase (hbd-9,2 af2017) from archaeoglobus fulgidus dsm 4304 at 2.00 a resolution
Phyre2