1 c1qmiC_
100.0
99
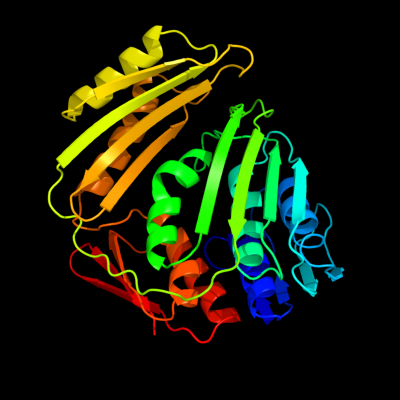
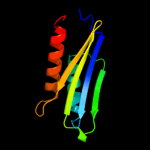
PDB header: rna 3'-terminal phosphate cyclaseChain: C: PDB Molecule: rna 3'-terminal phosphate cyclase;PDBTitle: crystal structure of rna 3'-terminal phosphate cyclase, an2 ubiquitous enzyme with unusual topology
2 c3pqvD_
100.0
18
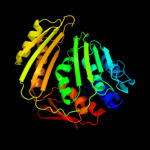
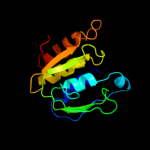
PDB header: unknown functionChain: D: PDB Molecule: rcl1 protein;PDBTitle: cyclase homolog
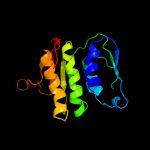
3 d1qmha2
100.0
100
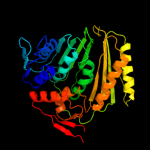
Fold: IF3-likeSuperfamily: EPT/RTPC-likeFamily: RNA 3'-terminal phosphate cyclase, RPTC4 d1qmha1
99.9
100
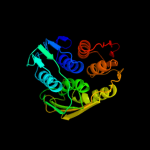
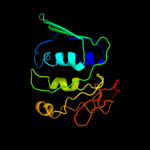
Fold: Thioredoxin foldSuperfamily: RNA 3'-terminal phosphate cyclase, RPTC, insert domainFamily: RNA 3'-terminal phosphate cyclase, RPTC, insert domain5 d1p88a_
95.7
15
Fold: IF3-likeSuperfamily: EPT/RTPC-likeFamily: Enolpyruvate transferase, EPT6 c2o0zA_
94.7
20
PDB header: transferaseChain: A: PDB Molecule: 3-phosphoshikimate 1-carboxyvinyltransferase;PDBTitle: mycobacterium tuberculosis epsp synthase in complex with2 product (eps)
7 d1uaea_
94.5
17
Fold: IF3-likeSuperfamily: EPT/RTPC-likeFamily: Enolpyruvate transferase, EPT8 d1g6sa_
93.0
21
Fold: IF3-likeSuperfamily: EPT/RTPC-likeFamily: Enolpyruvate transferase, EPT9 c2pqdA_
92.0
17
PDB header: transferaseChain: A: PDB Molecule: 3-phosphoshikimate 1-carboxyvinyltransferase;PDBTitle: a100g cp4 epsps liganded with (r)-difluoromethyl tetrahedral reaction2 intermediate analog
10 c3roiA_
91.8
21
PDB header: transferaseChain: A: PDB Molecule: 3-phosphoshikimate 1-carboxyvinyltransferase;PDBTitle: 2.20 angstrom resolution structure of 3-phosphoshikimate 1-2 carboxyvinyltransferase (aroa) from coxiella burnetii
11 c3rmtB_
91.7
21
PDB header: transferaseChain: B: PDB Molecule: 3-phosphoshikimate 1-carboxyvinyltransferase 1;PDBTitle: crystal structure of putative 5-enolpyruvoylshikimate-3-phosphate2 synthase from bacillus halodurans c-125
12 c2yvwA_
87.1
21
PDB header: transferaseChain: A: PDB Molecule: udp-n-acetylglucosamine 1-carboxyvinyltransferase;PDBTitle: crystal structure of udp-n-acetylglucosamine 1-carboxyvinyltransferase2 from aquifex aeolicus vf5
13 d1ejda_
81.4
17
Fold: IF3-likeSuperfamily: EPT/RTPC-likeFamily: Enolpyruvate transferase, EPT14 c3r38A_
73.0
11
PDB header: transferaseChain: A: PDB Molecule: udp-n-acetylglucosamine 1-carboxyvinyltransferase 1;PDBTitle: 2.23 angstrom resolution crystal structure of udp-n-acetylglucosamine2 1-carboxyvinyltransferase (mura) from listeria monocytogenes egd-e
15 d1rf6a_
69.6
13
Fold: IF3-likeSuperfamily: EPT/RTPC-likeFamily: Enolpyruvate transferase, EPT16 d1ul7a_
46.3
14
Fold: TBP-likeSuperfamily: KA1-likeFamily: Kinase associated domain 1, KA117 c3cvoA_
34.5
21
PDB header: transferaseChain: A: PDB Molecule: methyltransferase-like protein of unknown function;PDBTitle: crystal structure of a methyltransferase-like protein (spo2022) from2 silicibacter pomeroyi dss-3 at 1.80 a resolution
18 c2uwjE_
34.3
24
PDB header: chaperoneChain: E: PDB Molecule: type iii export protein psce;PDBTitle: structure of the heterotrimeric complex which regulates2 type iii secretion needle formation
19 c1hskA_
28.2
23
PDB header: oxidoreductaseChain: A: PDB Molecule: udp-n-acetylenolpyruvoylglucosamine reductase;PDBTitle: crystal structure of s. aureus murb
20 d2e1ba1
26.7
75
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: AlaX-M N-terminal domain-like21 d1hska2
not modelled
23.6
23
Fold: Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase, MurB, C-terminal domainSuperfamily: Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase, MurB, C-terminal domainFamily: Uridine diphospho-N-Acetylenolpyruvylglucosamine reductase, MurB, C-terminal domain22 d1f46a_
not modelled
22.2
12
Fold: TBP-likeSuperfamily: Cell-division protein ZipA, C-terminal domainFamily: Cell-division protein ZipA, C-terminal domain23 c2e1bA_
not modelled
16.0
75
PDB header: ligase, hydrolaseChain: A: PDB Molecule: 216aa long hypothetical alanyl-trna synthetase;PDBTitle: crystal structure of the alax-m trans-editing enzyme from2 pyrococcus horikoshii
24 c2q1kA_
not modelled
13.2
29
PDB header: chaperoneChain: A: PDB Molecule: asce;PDBTitle: cyrstal structure of asce from aeromonas hydrophilla
25 d2q07a3
not modelled
11.4
38
Fold: Cystatin-likeSuperfamily: Pre-PUA domainFamily: AF0587 pre C-terminal domain-like26 d1h72c1
not modelled
10.9
16
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: GHMP Kinase, N-terminal domain27 d2od4a1
not modelled
10.9
21
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: Marine metagenome family DABB228 d1mp9a1
not modelled
10.7
23
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain29 d1hh2p3
not modelled
9.9
5
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Prokaryotic type KH domain (KH-domain type II)Family: Prokaryotic type KH domain (KH-domain type II)30 d2zdra1
not modelled
9.8
12
Fold: beta-clipSuperfamily: AFP III-like domainFamily: AFP III-like domain31 d1nh2a1
not modelled
9.4
28
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain32 c2xi5D_
not modelled
9.4
13
PDB header: transferaseChain: D: PDB Molecule: rna polymerase l;PDBTitle: n-terminal endonuclease domain of la crosse virus l-protein
33 c2xi7B_
not modelled
9.3
13
PDB header: transferaseChain: B: PDB Molecule: rna polymerase l;PDBTitle: n-terminal endonuclease domain of la crosse virus l-protein
34 d2a9sa1
not modelled
9.3
5
Fold: Anticodon-binding domain-likeSuperfamily: CinA-likeFamily: CinA-like35 d1ul1x1
not modelled
9.1
12
Fold: SAM domain-likeSuperfamily: 5' to 3' exonuclease, C-terminal subdomainFamily: 5' to 3' exonuclease, C-terminal subdomain36 d1nwpa_
not modelled
9.0
21
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like37 d1cuoa_
not modelled
8.3
13
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like38 c2ch4A_
not modelled
7.8
18
PDB header: transferase/chemotaxisChain: A: PDB Molecule: chemotaxis protein chea;PDBTitle: complex between bacterial chemotaxis histidine kinase chea2 domains p4 and p5 and receptor-adaptor protein chew
39 d1cf2o2
not modelled
7.4
36
Fold: FwdE/GAPDH domain-likeSuperfamily: Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domainFamily: GAPDH-like40 d1rkra_
not modelled
7.3
21
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like41 d1qnaa1
not modelled
7.3
20
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain42 c1fwlD_
not modelled
7.0
16
PDB header: transferaseChain: D: PDB Molecule: homoserine kinase;PDBTitle: crystal structure of homoserine kinase
43 d1aisa1
not modelled
6.7
26
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain44 c1gmuB_
not modelled
6.6
31
PDB header: metallochaperoneChain: B: PDB Molecule: uree;PDBTitle: structure of uree
45 d1cc3a_
not modelled
6.4
17
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Plastocyanin/azurin-like46 d1mp9a2
not modelled
5.8
14
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain47 c2rrnA_
not modelled
5.8
14
PDB header: protein transportChain: A: PDB Molecule: probable secdf protein-export membrane protein;PDBTitle: solution structure of secdf periplasmic domain p4
48 d1cdwa1
not modelled
5.4
18
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: TATA-box binding protein (TBP), C-terminal domain49 d1gmua2
not modelled
5.3
30
Fold: Ferredoxin-likeSuperfamily: Urease metallochaperone UreE, C-terminal domainFamily: Urease metallochaperone UreE, C-terminal domain