| 1 |
|
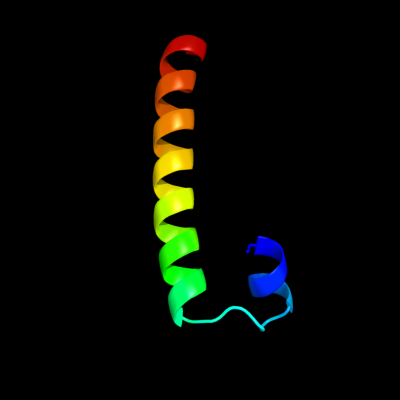
PDB 3bhp chain A
Region: 39 - 75
Aligned: 37
Modelled: 37
Confidence: 40.4%
Identity: 19%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0291 protein ynzc;
PDBTitle: crystal structure of upf0291 protein ynzc from bacillus2 subtilis at resolution 2.0 a. northeast structural3 genomics consortium target sr384
Phyre2
| 2 |
|
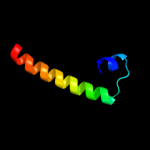
PDB 1t08 chain B
Region: 44 - 79
Aligned: 36
Modelled: 36
Confidence: 32.0%
Identity: 36%
Fold: beta-catenin-interacting protein ICAT
Superfamily: beta-catenin-interacting protein ICAT
Family: beta-catenin-interacting protein ICAT
Phyre2
| 3 |
|
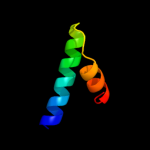
PDB 1m1e chain B
Region: 44 - 85
Aligned: 42
Modelled: 42
Confidence: 25.8%
Identity: 33%
Fold: beta-catenin-interacting protein ICAT
Superfamily: beta-catenin-interacting protein ICAT
Family: beta-catenin-interacting protein ICAT
Phyre2
| 4 |
|
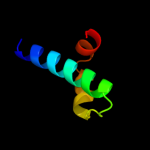
PDB 1st6 chain A domain 5
Region: 13 - 88
Aligned: 76
Modelled: 76
Confidence: 18.4%
Identity: 16%
Fold: Four-helical up-and-down bundle
Superfamily: alpha-catenin/vinculin-like
Family: alpha-catenin/vinculin
Phyre2
| 5 |
|
PDB 1luj chain B
Region: 44 - 79
Aligned: 36
Modelled: 36
Confidence: 18.2%
Identity: 36%
Fold: beta-catenin-interacting protein ICAT
Superfamily: beta-catenin-interacting protein ICAT
Family: beta-catenin-interacting protein ICAT
Phyre2
| 6 |
|
PDB 2vn2 chain B
Region: 65 - 84
Aligned: 20
Modelled: 20
Confidence: 13.8%
Identity: 40%
PDB header:replication
Chain: B: PDB Molecule:chromosome replication initiation protein;
PDBTitle: crystal structure of the n-terminal domain of dnad protein2 from geobacillus kaustophilus hta426
Phyre2
| 7 |
|
PDB 1uix chain A
Region: 22 - 82
Aligned: 59
Modelled: 61
Confidence: 13.4%
Identity: 24%
PDB header:transferase
Chain: A: PDB Molecule:rho-associated kinase;
PDBTitle: coiled-coil structure of the rhoa-binding domain in rho-2 kinase
Phyre2
| 8 |
|
PDB 3lay chain F
Region: 48 - 93
Aligned: 46
Modelled: 46
Confidence: 13.1%
Identity: 20%
PDB header:metal binding protein
Chain: F: PDB Molecule:zinc resistance-associated protein;
PDBTitle: alpha-helical barrel formed by the decamer of the zinc resistance-2 associated protein (stm4172) from salmonella enterica subsp. enterica3 serovar typhimurium str. lt2
Phyre2
| 9 |
|
PDB 2jvd chain A
Region: 39 - 66
Aligned: 28
Modelled: 28
Confidence: 11.1%
Identity: 21%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0291 protein ynzc;
PDBTitle: solution nmr structure of the folded n-terminal fragment of2 upf0291 protein ynzc from bacillus subtilis. northeast3 structural genomics target sr384-1-46
Phyre2
| 10 |
|
PDB 2fup chain A domain 1
Region: 23 - 77
Aligned: 52
Modelled: 55
Confidence: 10.9%
Identity: 13%
Fold: STAT-like
Superfamily: FlgN-like
Family: FlgN-like
Phyre2
| 11 |
|
PDB 2fup chain A
Region: 23 - 77
Aligned: 52
Modelled: 55
Confidence: 10.9%
Identity: 13%
PDB header:biosynthetic protein
Chain: A: PDB Molecule:hypothetical protein pa3352;
PDBTitle: crystal structure of a putative flagella synthesis protein flgn2 (pa3352) from pseudomonas aeruginosa at 1.48 a resolution
Phyre2
| 12 |
|
PDB 1rh4 chain A
Region: 52 - 69
Aligned: 18
Modelled: 18
Confidence: 7.8%
Identity: 56%
PDB header:coiled coil
Chain: A: PDB Molecule:right-handed coiled coil tetramer;
PDBTitle: rh4 designed right-handed coiled coil tetramer
Phyre2
| 13 |
|
PDB 3qsq chain A
Region: 9 - 21
Aligned: 13
Modelled: 13
Confidence: 7.0%
Identity: 38%
PDB header:viral protein
Chain: A: PDB Molecule:capsid polyprotein;
PDBTitle: crystal structure of the projection domain of the human astrovirus2 capsid protein
Phyre2
| 14 |
|
PDB 1hbw chain A
Region: 64 - 76
Aligned: 13
Modelled: 13
Confidence: 7.0%
Identity: 46%
PDB header:transcriptional activator
Chain: A: PDB Molecule:regulatory protein gal4;
PDBTitle: solution nmr structure of the dimerization domain of the2 yeast transcriptional activator gal4 (residues 50-106)
Phyre2
| 15 |
|
PDB 1s1h chain O
Region: 34 - 64
Aligned: 31
Modelled: 31
Confidence: 6.9%
Identity: 16%
PDB header:ribosome
Chain: O: PDB Molecule:40s ribosomal protein s13;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1h,4 contains 40s subunit. the 60s ribosomal subunit is in file5 1s1i.
Phyre2
| 16 |
|
PDB 1nb4 chain A
Region: 44 - 97
Aligned: 54
Modelled: 54
Confidence: 5.7%
Identity: 24%
Fold: DNA/RNA polymerases
Superfamily: DNA/RNA polymerases
Family: RNA-dependent RNA-polymerase
Phyre2
| 17 |
|
PDB 3aap chain A
Region: 56 - 108
Aligned: 53
Modelled: 53
Confidence: 5.5%
Identity: 26%
PDB header:hydrolase
Chain: A: PDB Molecule:ectonucleoside triphosphate diphosphohydrolase i;
PDBTitle: crystal structure of lp1ntpdase from legionella pneumophila
Phyre2