| 1 |
|
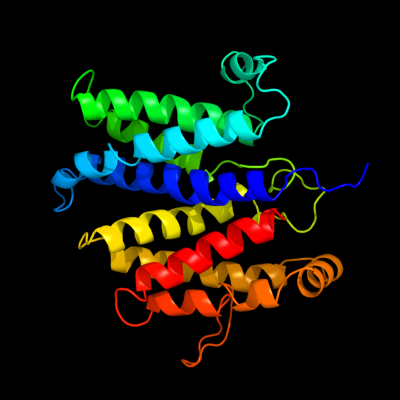
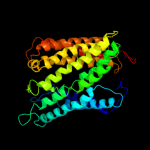
PDB 2vpz chain G
Region: 46 - 332
Aligned: 236
Modelled: 255
Confidence: 100.0%
Identity: 17%
PDB header:oxidoreductase
Chain: G: PDB Molecule:hypothetical membrane spanning protein;
PDBTitle: polysulfide reductase native structure
Phyre2
| 2 |
|
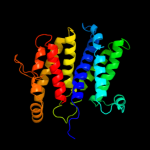
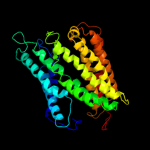
PDB 1ar1 chain A
Region: 5 - 390
Aligned: 375
Modelled: 386
Confidence: 89.6%
Identity: 12%
Fold: Cytochrome c oxidase subunit I-like
Superfamily: Cytochrome c oxidase subunit I-like
Family: Cytochrome c oxidase subunit I-like
Phyre2
| 3 |
|
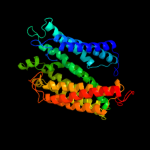
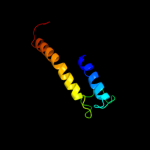
PDB 1v54 chain A
Region: 5 - 386
Aligned: 372
Modelled: 381
Confidence: 62.2%
Identity: 11%
Fold: Cytochrome c oxidase subunit I-like
Superfamily: Cytochrome c oxidase subunit I-like
Family: Cytochrome c oxidase subunit I-like
Phyre2
| 4 |
|
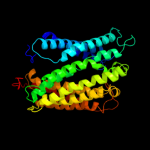
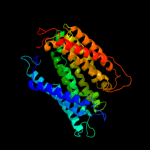
PDB 1fft chain A
Region: 5 - 388
Aligned: 374
Modelled: 375
Confidence: 61.6%
Identity: 11%
Fold: Cytochrome c oxidase subunit I-like
Superfamily: Cytochrome c oxidase subunit I-like
Family: Cytochrome c oxidase subunit I-like
Phyre2
| 5 |
|
PDB 1fft chain F
Region: 5 - 388
Aligned: 374
Modelled: 375
Confidence: 61.6%
Identity: 11%
PDB header:oxidoreductase
Chain: F: PDB Molecule:ubiquinol oxidase;
PDBTitle: the structure of ubiquinol oxidase from escherichia coli
Phyre2
| 6 |
|
PDB 3qnq chain D
Region: 313 - 388
Aligned: 76
Modelled: 76
Confidence: 48.5%
Identity: 4%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 7 |
|
PDB 1m56 chain G
Region: 5 - 388
Aligned: 373
Modelled: 384
Confidence: 39.0%
Identity: 12%
PDB header:oxidoreductase
Chain: G: PDB Molecule:cytochrome c oxidase;
PDBTitle: structure of cytochrome c oxidase from rhodobactor2 sphaeroides (wild type)
Phyre2
| 8 |
|
PDB 2b2h chain A
Region: 218 - 391
Aligned: 173
Modelled: 174
Confidence: 10.5%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter;
PDBTitle: ammonium transporter amt-1 from a. fulgidus (as)
Phyre2
| 9 |
|
PDB 2bbj chain B
Region: 14 - 83
Aligned: 55
Modelled: 70
Confidence: 9.5%
Identity: 20%
PDB header:metal transport/membrane protein
Chain: B: PDB Molecule:divalent cation transport-related protein;
PDBTitle: crystal structure of the cora mg2+ transporter
Phyre2
| 10 |
|
PDB 1vcn chain A
Region: 108 - 129
Aligned: 22
Modelled: 22
Confidence: 7.7%
Identity: 18%
PDB header:ligase
Chain: A: PDB Molecule:ctp synthetase;
PDBTitle: crystal structure of t.th. hb8 ctp synthetase complex with sulfate2 anion
Phyre2
| 11 |
|
PDB 1vco chain A domain 2
Region: 108 - 129
Aligned: 22
Modelled: 22
Confidence: 7.3%
Identity: 18%
Fold: P-loop containing nucleoside triphosphate hydrolases
Superfamily: P-loop containing nucleoside triphosphate hydrolases
Family: Nitrogenase iron protein-like
Phyre2
| 12 |
|
PDB 1s1m chain A domain 2
Region: 108 - 129
Aligned: 22
Modelled: 22
Confidence: 6.9%
Identity: 18%
Fold: P-loop containing nucleoside triphosphate hydrolases
Superfamily: P-loop containing nucleoside triphosphate hydrolases
Family: Nitrogenase iron protein-like
Phyre2
| 13 |
|
PDB 3nva chain B
Region: 108 - 129
Aligned: 22
Modelled: 22
Confidence: 6.9%
Identity: 18%
PDB header:ligase
Chain: B: PDB Molecule:ctp synthase;
PDBTitle: dimeric form of ctp synthase from sulfolobus solfataricus
Phyre2
| 14 |
|
PDB 1kp0 chain A domain 1
Region: 108 - 115
Aligned: 8
Modelled: 8
Confidence: 6.6%
Identity: 50%
Fold: Ribonuclease H-like motif
Superfamily: Creatinase/prolidase N-terminal domain
Family: Creatinase/prolidase N-terminal domain
Phyre2
| 15 |
|
PDB 3j01 chain A
Region: 218 - 392
Aligned: 174
Modelled: 175
Confidence: 5.8%
Identity: 9%
PDB header:ribosome/ribosomal protein
Chain: A: PDB Molecule:preprotein translocase secy subunit;
PDBTitle: structure of the ribosome-secye complex in the membrane environment
Phyre2