| 1 |
|
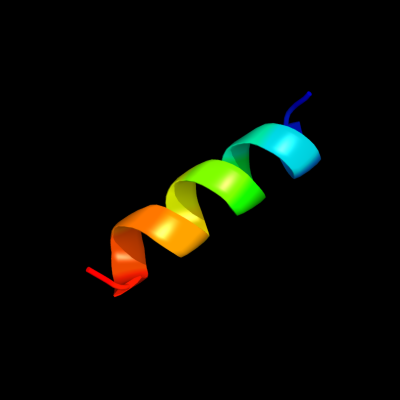
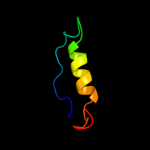
PDB 1eys chain H domain 2
Region: 91 - 105
Aligned: 15
Modelled: 15
Confidence: 40.7%
Identity: 20%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction centre subunit H, transmembrane region
Family: Photosystem II reaction centre subunit H, transmembrane region
Phyre2
| 2 |
|
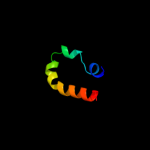
PDB 1eys chain H
Region: 91 - 105
Aligned: 15
Modelled: 15
Confidence: 29.2%
Identity: 20%
PDB header:electron transport
Chain: H: PDB Molecule:photosynthetic reaction center;
PDBTitle: crystal structure of photosynthetic reaction center from a2 thermophilic bacterium, thermochromatium tepidum
Phyre2
| 3 |
|
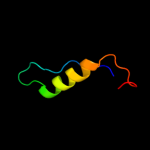
PDB 2i5n chain H
Region: 89 - 105
Aligned: 17
Modelled: 17
Confidence: 28.9%
Identity: 18%
PDB header:photosynthesis
Chain: H: PDB Molecule:reaction center protein h chain;
PDBTitle: 1.96 a x-ray structure of photosynthetic reaction center from2 rhodopseudomonas viridis:crystals grown by microfluidic technique
Phyre2
| 4 |
|
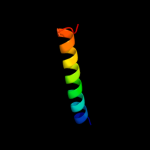
PDB 1k6n chain H
Region: 91 - 105
Aligned: 15
Modelled: 15
Confidence: 28.3%
Identity: 27%
PDB header:photosynthesis
Chain: H: PDB Molecule:photosynthetic reaction center h subunit;
PDBTitle: e(l212)a,d(l213)a double mutant structure of photosynthetic reaction2 center from rhodobacter sphaeroides
Phyre2
| 5 |
|
PDB 2kic chain A
Region: 53 - 71
Aligned: 19
Modelled: 19
Confidence: 20.3%
Identity: 11%
PDB header:protein binding,metal transport
Chain: A: PDB Molecule:nitrogenase gamma subunit;
PDBTitle: n-nafy. n-terminal domain of nafy
Phyre2
| 6 |
|
PDB 1hb6 chain A
Region: 31 - 67
Aligned: 33
Modelled: 37
Confidence: 16.3%
Identity: 18%
Fold: Acyl-CoA binding protein-like
Superfamily: Acyl-CoA binding protein
Family: Acyl-CoA binding protein
Phyre2
| 7 |
|
PDB 3kwr chain A
Region: 17 - 37
Aligned: 21
Modelled: 21
Confidence: 13.0%
Identity: 33%
PDB header:rna binding protein
Chain: A: PDB Molecule:putative rna-binding protein;
PDBTitle: crystal structure of putative rna-binding protein (np_785364.1) from2 lactobacillus plantarum at 1.45 a resolution
Phyre2
| 8 |
|
PDB 2elo chain A
Region: 60 - 80
Aligned: 21
Modelled: 21
Confidence: 12.9%
Identity: 10%
PDB header:transcription
Chain: A: PDB Molecule:zinc finger protein 406;
PDBTitle: solution structure of the 12th c2h2 zinc finger of human2 zinc finger protein 406
Phyre2
| 9 |
|
PDB 3epy chain A
Region: 31 - 67
Aligned: 33
Modelled: 37
Confidence: 12.5%
Identity: 18%
PDB header:lipid binding protein
Chain: A: PDB Molecule:acyl-coa-binding domain-containing protein 7;
PDBTitle: crystal structure of human acyl-coa binding domain 72 complexed with palmitoyl-coa
Phyre2
| 10 |
|
PDB 1st7 chain A
Region: 31 - 67
Aligned: 33
Modelled: 37
Confidence: 9.9%
Identity: 18%
PDB header:transport protein
Chain: A: PDB Molecule:acyl-coa-binding protein;
PDBTitle: solution structure of acyl coenzyme a binding protein from2 yeast
Phyre2
| 11 |
|
PDB 2cop chain A
Region: 31 - 67
Aligned: 33
Modelled: 37
Confidence: 9.3%
Identity: 18%
PDB header:lipid binding protein
Chain: A: PDB Molecule:acyl-coenzyme a binding domain containing 6;
PDBTitle: solution structure of rsgi ruh-040, an acbp domain from2 human cdna
Phyre2
| 12 |
|
PDB 2wh5 chain A
Region: 31 - 67
Aligned: 33
Modelled: 37
Confidence: 9.0%
Identity: 21%
PDB header:lipid binding protein
Chain: A: PDB Molecule:acyl-coa-binding domain-containing protein 4;
PDBTitle: crystal structure of human acyl-coa binding domain 42 complexed with stearoyl-coa
Phyre2
| 13 |
|
PDB 1hbk chain A
Region: 31 - 67
Aligned: 33
Modelled: 37
Confidence: 8.2%
Identity: 9%
Fold: Acyl-CoA binding protein-like
Superfamily: Acyl-CoA binding protein
Family: Acyl-CoA binding protein
Phyre2
| 14 |
|
PDB 4a1e chain E
Region: 42 - 62
Aligned: 19
Modelled: 21
Confidence: 7.8%
Identity: 21%
PDB header:ribosome
Chain: E: PDB Molecule:60s ribosomal protein l9;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna, 5.8s rrna3 and proteins of molecule 1
Phyre2
| 15 |
|
PDB 3iz5 chain F
Region: 42 - 62
Aligned: 19
Modelled: 21
Confidence: 7.1%
Identity: 21%
PDB header:ribosome
Chain: F: PDB Molecule:60s ribosomal protein l9 (l6p);
PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
Phyre2
| 16 |
|
PDB 2lbb chain A
Region: 31 - 67
Aligned: 33
Modelled: 37
Confidence: 6.9%
Identity: 15%
PDB header:protein binding
Chain: A: PDB Molecule:acyl coa binding protein;
PDBTitle: solution structure of acyl coa binding protein from babesia bovis t2bo
Phyre2
| 17 |
|
PDB 2iec chain A domain 1
Region: 22 - 60
Aligned: 29
Modelled: 39
Confidence: 6.2%
Identity: 24%
Fold: MK0786-like
Superfamily: MK0786-like
Family: MK0786-like
Phyre2
| 18 |
|
PDB 3fp5 chain A
Region: 31 - 67
Aligned: 33
Modelled: 37
Confidence: 6.1%
Identity: 18%
PDB header:lipid binding protein
Chain: A: PDB Molecule:acyl-coa binding protein;
PDBTitle: crystal structure of acbp from moniliophthora perniciosa
Phyre2
| 19 |
|
PDB 3g80 chain B
Region: 51 - 63
Aligned: 13
Modelled: 13
Confidence: 5.7%
Identity: 38%
PDB header:viral protein
Chain: B: PDB Molecule:protein b2;
PDBTitle: nodamura virus protein b2, rna-binding domain
Phyre2
| 20 |
|
PDB 1afo chain B
Region: 78 - 105
Aligned: 28
Modelled: 28
Confidence: 5.7%
Identity: 14%
PDB header:integral membrane protein
Chain: B: PDB Molecule:glycophorin a;
PDBTitle: dimeric transmembrane domain of human glycophorin a, nmr,2 20 structures
Phyre2
| 21 |
|