| 1 |
|
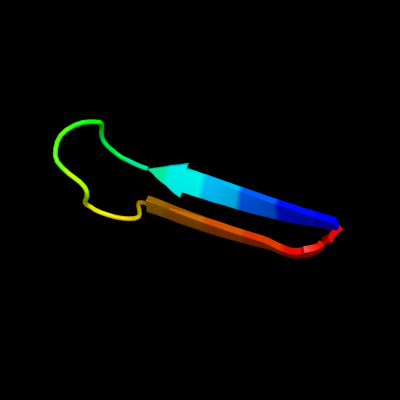
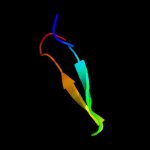
PDB 1f0c chain B
Region: 104 - 121
Aligned: 18
Modelled: 18
Confidence: 12.4%
Identity: 28%
PDB header:viral protein
Chain: B: PDB Molecule:ice inhibitor;
PDBTitle: structure of the viral serpin crma
Phyre2
| 2 |
|
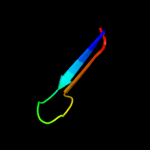
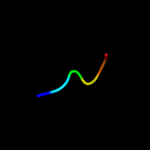
PDB 3c0n chain A domain 1
Region: 79 - 93
Aligned: 15
Modelled: 15
Confidence: 11.2%
Identity: 33%
Fold: C-type lectin-like
Superfamily: C-type lectin-like
Family: Aerolysin/Pertussis toxin (APT) domain
Phyre2
| 3 |
|
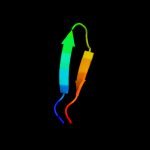
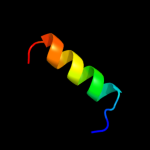
PDB 2vp7 chain B
Region: 49 - 67
Aligned: 19
Modelled: 19
Confidence: 10.6%
Identity: 26%
PDB header:gene regulation
Chain: B: PDB Molecule:b-cell cll/lymphoma 9 protein;
PDBTitle: decoding of methylated histone h3 tail by the pygo-bcl9 wnt2 signaling complex
Phyre2
| 4 |
|
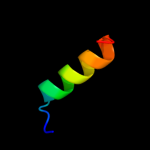
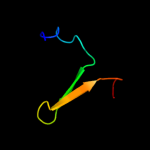
PDB 2oai chain A domain 1
Region: 76 - 94
Aligned: 19
Modelled: 19
Confidence: 10.3%
Identity: 21%
Fold: FAD-binding/transporter-associated domain-like
Superfamily: FAD-binding/transporter-associated domain-like
Family: CorC/HlyC domain-like
Phyre2
| 5 |
|
PDB 2yrm chain A
Region: 16 - 21
Aligned: 6
Modelled: 6
Confidence: 10.1%
Identity: 67%
PDB header:gene regulation
Chain: A: PDB Molecule:b-cell lymphoma 6 protein;
PDBTitle: solution structure of the 1st zf-c2h2 domain from human b-2 cell lymphoma 6 protein
Phyre2
| 6 |
|
PDB 2vpd chain B
Region: 49 - 67
Aligned: 19
Modelled: 19
Confidence: 10.0%
Identity: 26%
PDB header:gene regulation
Chain: B: PDB Molecule:b-cell cll/lymphoma 9 protein;
PDBTitle: decoding of methylated histone h3 tail by the pygo-bcl9 wnt2 signaling complex
Phyre2
| 7 |
|
PDB 1ept chain A
Region: 69 - 99
Aligned: 31
Modelled: 31
Confidence: 9.2%
Identity: 19%
PDB header:hydrolase (serine protease)
Chain: A: PDB Molecule:porcine e-trypsin;
PDBTitle: refined 1.8 angstroms resolution crystal structure of2 porcine epsilon-trypsin
Phyre2
| 8 |
|
PDB 2vpd chain D
Region: 49 - 67
Aligned: 19
Modelled: 19
Confidence: 9.0%
Identity: 26%
PDB header:gene regulation
Chain: D: PDB Molecule:b-cell cll/lymphoma 9 protein;
PDBTitle: decoding of methylated histone h3 tail by the pygo-bcl9 wnt2 signaling complex
Phyre2
| 9 |
|
PDB 2fa8 chain A domain 1
Region: 93 - 116
Aligned: 19
Modelled: 24
Confidence: 8.5%
Identity: 32%
Fold: Thioredoxin fold
Superfamily: Thioredoxin-like
Family: Selenoprotein W-related
Phyre2
| 10 |
|
PDB 2obk chain E
Region: 93 - 116
Aligned: 19
Modelled: 24
Confidence: 7.8%
Identity: 32%
PDB header:structural genomics, unknown function
Chain: E: PDB Molecule:selt/selw/selh selenoprotein domain;
PDBTitle: x-ray structure of the putative se binding protein from pseudomonas2 fluorescens. northeast structural genomics consortium target plr6.
Phyre2
| 11 |
|
PDB 3thd chain D
Region: 76 - 105
Aligned: 28
Modelled: 30
Confidence: 7.6%
Identity: 21%
PDB header:hydrolase
Chain: D: PDB Molecule:beta-galactosidase;
PDBTitle: crystal structure of human beta-galactosidase in complex with 1-2 deoxygalactonojirimycin
Phyre2
| 12 |
|
PDB 2p13 chain A domain 1
Region: 76 - 93
Aligned: 18
Modelled: 18
Confidence: 7.2%
Identity: 22%
Fold: FAD-binding/transporter-associated domain-like
Superfamily: FAD-binding/transporter-associated domain-like
Family: CorC/HlyC domain-like
Phyre2
| 13 |
|
PDB 2npb chain A
Region: 85 - 117
Aligned: 28
Modelled: 33
Confidence: 6.4%
Identity: 25%
PDB header:oxidoreductase
Chain: A: PDB Molecule:selenoprotein w;
PDBTitle: nmr solution structure of mouse selw
Phyre2