| 1 |
|
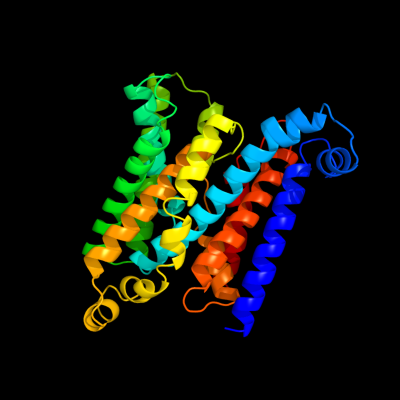
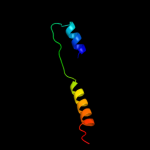
PDB 1l7v chain A
Region: 5 - 325
Aligned: 313
Modelled: 321
Confidence: 100.0%
Identity: 31%
Fold: ABC transporter involved in vitamin B12 uptake, BtuC
Superfamily: ABC transporter involved in vitamin B12 uptake, BtuC
Family: ABC transporter involved in vitamin B12 uptake, BtuC
Phyre2
| 2 |
|
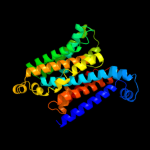
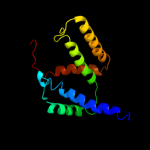
PDB 2nq2 chain A
Region: 8 - 326
Aligned: 306
Modelled: 319
Confidence: 100.0%
Identity: 27%
PDB header:metal transport
Chain: A: PDB Molecule:hypothetical abc transporter permease protein
PDBTitle: an inward-facing conformation of a putative metal-chelate2 type abc transporter.
Phyre2
| 3 |
|
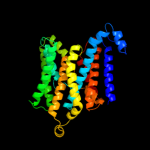
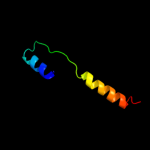
PDB 2b2h chain A
Region: 57 - 231
Aligned: 166
Modelled: 171
Confidence: 60.8%
Identity: 13%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter;
PDBTitle: ammonium transporter amt-1 from a. fulgidus (as)
Phyre2
| 4 |
|
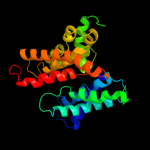
PDB 1yce chain D
Region: 75 - 116
Aligned: 42
Modelled: 42
Confidence: 16.2%
Identity: 19%
PDB header:membrane protein
Chain: D: PDB Molecule:subunit c;
PDBTitle: structure of the rotor ring of f-type na+-atpase from ilyobacter2 tartaricus
Phyre2
| 5 |
|
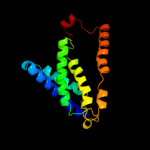
PDB 3lrc chain C
Region: 190 - 329
Aligned: 140
Modelled: 140
Confidence: 12.1%
Identity: 7%
PDB header:transport protein
Chain: C: PDB Molecule:arginine/agmatine antiporter;
PDBTitle: structure of e. coli adic (p1)
Phyre2
| 6 |
|
PDB 2w5j chain M
Region: 75 - 115
Aligned: 41
Modelled: 41
Confidence: 10.0%
Identity: 20%
PDB header:hydrolase
Chain: M: PDB Molecule:atp synthase c chain, chloroplastic;
PDBTitle: structure of the c14-rotor ring of the proton translocating2 chloroplast atp synthase
Phyre2
| 7 |
|
PDB 1q1h chain A
Region: 220 - 241
Aligned: 22
Modelled: 22
Confidence: 9.9%
Identity: 23%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: "Winged helix" DNA-binding domain
Family: Transcription factor E/IIe-alpha, N-terminal domain
Phyre2
| 8 |
|
PDB 1q1h chain A
Region: 220 - 241
Aligned: 22
Modelled: 22
Confidence: 9.9%
Identity: 23%
PDB header:transcription
Chain: A: PDB Molecule:transcription factor e;
PDBTitle: an extended winged helix domain in general transcription2 factor e/iie alpha
Phyre2
| 9 |
|
PDB 3k3g chain A
Region: 61 - 330
Aligned: 249
Modelled: 253
Confidence: 8.9%
Identity: 15%
PDB header:transport protein
Chain: A: PDB Molecule:urea transporter;
PDBTitle: crystal structure of the urea transporter from desulfovibrio vulgaris2 bound to 1,3-dimethylurea
Phyre2
| 10 |
|
PDB 2xq2 chain A
Region: 11 - 210
Aligned: 183
Modelled: 193
Confidence: 6.0%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: structure of the k294a mutant of vsglt
Phyre2