| 1 |
|
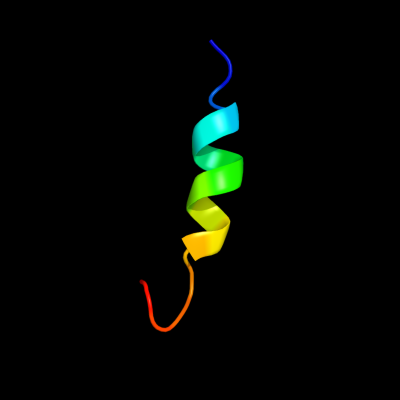
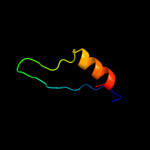
PDB 2dii chain A
Region: 45 - 61
Aligned: 17
Modelled: 17
Confidence: 28.2%
Identity: 47%
PDB header:transcription
Chain: A: PDB Molecule:tfiih basal transcription factor complex p62
PDBTitle: solution structure of the bsd domain of human tfiih basal2 transcription factor complex p62 subunit
Phyre2
| 2 |
|
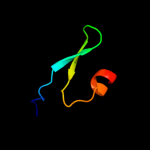
PDB 3p9v chain A
Region: 32 - 49
Aligned: 18
Modelled: 18
Confidence: 27.3%
Identity: 22%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: high resolution crystal structure of protein maqu_3174 from2 marinobacter aquaeolei, northeast structural genomics consortium3 target mqr197
Phyre2
| 3 |
|
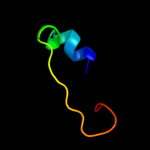
PDB 2dii chain A domain 1
Region: 45 - 61
Aligned: 17
Modelled: 17
Confidence: 24.7%
Identity: 47%
Fold: BSD domain-like
Superfamily: BSD domain-like
Family: BSD domain
Phyre2
| 4 |
|
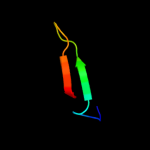
PDB 1zu1 chain A domain 1
Region: 48 - 61
Aligned: 14
Modelled: 14
Confidence: 23.4%
Identity: 29%
Fold: beta-beta-alpha zinc fingers
Superfamily: beta-beta-alpha zinc fingers
Family: HkH motif-containing C2H2 finger
Phyre2
| 5 |
|
PDB 2qwu chain B
Region: 30 - 57
Aligned: 28
Modelled: 28
Confidence: 12.1%
Identity: 25%
PDB header:cell invasion
Chain: B: PDB Molecule:intracellular growth locus, subunit c;
PDBTitle: crystal structure of f. tularensis pathogenicity island2 protein c
Phyre2
| 6 |
|
PDB 1y3n chain A domain 1
Region: 26 - 60
Aligned: 35
Modelled: 35
Confidence: 11.5%
Identity: 9%
Fold: Periplasmic binding protein-like II
Superfamily: Periplasmic binding protein-like II
Family: Phosphate binding protein-like
Phyre2
| 7 |
|
PDB 1k78 chain A domain 2
Region: 46 - 61
Aligned: 16
Modelled: 16
Confidence: 8.2%
Identity: 13%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: Paired domain
Phyre2
| 8 |
|
PDB 2l76 chain A
Region: 25 - 56
Aligned: 32
Modelled: 32
Confidence: 7.4%
Identity: 6%
PDB header:transcription
Chain: A: PDB Molecule:nfatc2-interacting protein;
PDBTitle: solution nmr structure of human nfatc2ip ubiquitin-like domain,2 nfatc2ip_244_338, nesg target ht65a/ocsp target hs00387_244_338/sgc-3 toronto
Phyre2
| 9 |
|
PDB 1p42 chain A domain 1
Region: 5 - 30
Aligned: 24
Modelled: 26
Confidence: 6.5%
Identity: 21%
Fold: Ribosomal protein S5 domain 2-like
Superfamily: Ribosomal protein S5 domain 2-like
Family: UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase LpxC
Phyre2
| 10 |
|
PDB 3i38 chain B
Region: 13 - 34
Aligned: 20
Modelled: 22
Confidence: 6.5%
Identity: 25%
PDB header:chaperone
Chain: B: PDB Molecule:putative chaperone dnaj;
PDBTitle: structure of a putative chaperone protein dnaj from klebsiella2 pneumoniae subsp. pneumoniae mgh 78578
Phyre2
| 11 |
|
PDB 6pax chain A domain 2
Region: 46 - 61
Aligned: 16
Modelled: 16
Confidence: 6.0%
Identity: 6%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: Paired domain
Phyre2
| 12 |
|
PDB 2o7h chain F
Region: 45 - 60
Aligned: 16
Modelled: 16
Confidence: 5.4%
Identity: 38%
PDB header:transcription
Chain: F: PDB Molecule:general control protein gcn4;
PDBTitle: crystal structure of trimeric coiled coil gcn4 leucine zipper
Phyre2