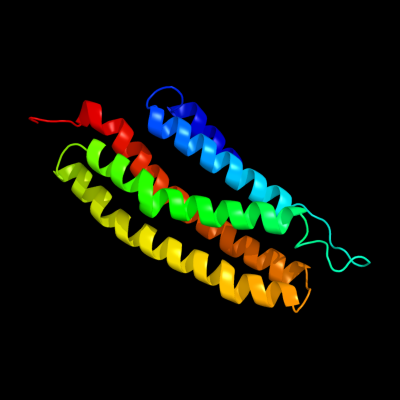
1 c2p7vA_
100.0
100
PDB header: transcriptionChain: A: PDB Molecule: regulator of sigma d;PDBTitle: crystal structure of the escherichia coli regulator of sigma 70, rsd,2 in complex with sigma 70 domain 4
2 c2wvmA_
44.1
19
PDB header: transferaseChain: A: PDB Molecule: mannosyl-3-phosphoglycerate synthase;PDBTitle: h309a mutant of mannosyl-3-phosphoglycerate synthase from2 thermus thermophilus hb27 in complex with3 gdp-alpha-d-mannose and mg(ii)
3 d1p94a_
39.8
55
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: CopG-like4 c2plyB_
30.5
19
PDB header: translation/rnaChain: B: PDB Molecule: selenocysteine-specific elongation factor;PDBTitle: structure of the mrna binding fragment of elongation factor2 selb in complex with secis rna.
5 c2zu8A_
28.9
8
PDB header: transferaseChain: A: PDB Molecule: mannosyl-3-phosphoglycerate synthase;PDBTitle: crystal structure of mannosyl-3-phosphoglycerate synthase2 from pyrococcus horikoshii
6 d1tlha_
28.4
16
Fold: Anti-sigma factor AsiASuperfamily: Anti-sigma factor AsiAFamily: Anti-sigma factor AsiA7 c3k1tA_
28.1
33
PDB header: ligaseChain: A: PDB Molecule: glutamate--cysteine ligase gsha;PDBTitle: crystal structure of putative gamma-glutamylcysteine synthetase2 (yp_546622.1) from methylobacillus flagellatus kt at 1.90 a3 resolution
8 c1wsuA_
27.8
19
PDB header: translation/rnaChain: A: PDB Molecule: selenocysteine-specific elongation factor;PDBTitle: c-terminal domain of elongation factor selb complexed with2 secis rna
9 c3hosA_
25.1
14
PDB header: transferase, dna binding protein/dnaChain: A: PDB Molecule: transposable element mariner, complete cds;PDBTitle: crystal structure of the mariner mos1 paired end complex with mg
10 c1avoD_
21.9
10
PDB header: proteasome activatorChain: D: PDB Molecule: 11s regulator;PDBTitle: proteasome activator reg(alpha)
11 d1uoua1
20.7
12
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain12 d2o3aa1
19.0
21
Fold: alpha/beta knotSuperfamily: alpha/beta knotFamily: AF0751-like13 c2yy8B_
17.3
14
PDB header: transferaseChain: B: PDB Molecule: upf0106 protein ph0461;PDBTitle: crystal structure of archaeal trna-methylase for position2 56 (atrm56) from pyrococcus horikoshii, complexed with s-3 adenosyl-l-methionine
14 d1tjla1
17.0
31
Fold: Long alpha-hairpinSuperfamily: DnaK suppressor protein DksA, alpha-hairpin domainFamily: DnaK suppressor protein DksA, alpha-hairpin domain15 d2tpta1
15.1
8
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain16 d1v92a_
11.4
17
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: TAP-C domain-like17 c2nrjA_
11.1
16
PDB header: toxinChain: A: PDB Molecule: hbl b protein;PDBTitle: crystal structure of hemolysin binding component from2 bacillus cereus
18 d1y2oa1
10.8
16
Fold: BAR/IMD domain-likeSuperfamily: BAR/IMD domain-likeFamily: IMD domain19 c2g8yB_
9.8
46
PDB header: oxidoreductaseChain: B: PDB Molecule: malate/l-lactate dehydrogenases;PDBTitle: the structure of a putative malate/lactate dehydrogenase from e. coli.
20 d2e9xa1
8.9
17
Fold: GINS helical bundle-likeSuperfamily: GINS helical bundle-likeFamily: PSF1 N-terminal domain-like21 d1d4ua1
not modelled
8.8
12
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: DNA repair factor XPA DNA- and RPA-binding domain, C-terminal subdomain22 c3pcsB_
not modelled
8.4
29
PDB header: protein transport/transferaseChain: B: PDB Molecule: espg;PDBTitle: structure of espg-pak2 autoinhibitory ialpha3 helix complex
23 d2fbqa2
not modelled
8.0
20
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain24 c1z56B_
not modelled
7.8
30
PDB header: ligaseChain: B: PDB Molecule: ligase interacting factor 1;PDBTitle: co-crystal structure of lif1p-lig4p
25 c3oqlB_
not modelled
7.5
9
PDB header: transcriptionChain: B: PDB Molecule: tena homolog;PDBTitle: crystal structure of a tena homolog (pspto1738) from pseudomonas2 syringae pv. tomato str. dc3000 at 2.54 a resolution
26 d2nvna1
not modelled
7.4
29
Fold: ssDNA-binding transcriptional regulator domainSuperfamily: ssDNA-binding transcriptional regulator domainFamily: PMN2A0962/syc2379c-like27 d1x2ma1
not modelled
7.3
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Homeodomain28 d1brwa1
not modelled
7.2
11
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain29 c3r89A_
not modelled
7.1
15
PDB header: lyaseChain: A: PDB Molecule: orotidine 5'-phosphate decarboxylase;PDBTitle: crystal structure of orotidine 5-phosphate decarboxylase from2 anaerococcus prevotii dsm 20548
30 c3dkaA_
not modelled
6.8
12
PDB header: unknown functionChain: A: PDB Molecule: dinb-like protein;PDBTitle: crystal structure of a dinb-like protein (yjoa, bsu12410) from2 bacillus subtilis at 2.30 a resolution
31 d2it9a1
not modelled
6.8
11
Fold: ssDNA-binding transcriptional regulator domainSuperfamily: ssDNA-binding transcriptional regulator domainFamily: PMN2A0962/syc2379c-like32 c2e19A_
not modelled
6.7
15
PDB header: transcriptionChain: A: PDB Molecule: transcription factor 8;PDBTitle: solution structure of the homeobox domain from human nil-2-2 a zinc finger protein, transcription factor 8
33 c3m9hB_
not modelled
6.6
28
PDB header: chaperoneChain: B: PDB Molecule: proteasome-associated atpase;PDBTitle: crystal structure of the amino terminal coiled coil domain of the2 mycobacterium tuberculosis proteasomal atpase mpa
34 d1v8ga1
not modelled
6.0
10
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain35 c1xawA_
not modelled
6.0
16
PDB header: cell adhesionChain: A: PDB Molecule: occludin;PDBTitle: crystal structure of the cytoplasmic distal c-terminal2 domain of occludin
36 c2q9qC_
not modelled
5.9
17
PDB header: replicationChain: C: PDB Molecule: dna replication complex gins protein psf1;PDBTitle: the crystal structure of full length human gins complex
37 c3e5aB_
not modelled
5.9
27
PDB header: transferaseChain: B: PDB Molecule: targeting protein for xklp2;PDBTitle: crystal structure of aurora a in complex with vx-680 and tpx2
38 d1z3xa2
not modelled
5.9
18
Fold: GUN4-likeSuperfamily: GUN4-likeFamily: GUN4-like39 d1oaia_
not modelled
5.7
17
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: TAP-C domain-like40 c2da7A_
not modelled
5.7
23
PDB header: dna binding proteinChain: A: PDB Molecule: zinc finger homeobox protein 1b;PDBTitle: solution structure of the homeobox domain of zinc finger2 homeobox protein 1b (smad interacting protein 1)
41 c1a92B_
not modelled
5.5
31
PDB header: leucine zipperChain: B: PDB Molecule: delta antigen;PDBTitle: oligomerization domain of hepatitis delta antigen
42 d2vo1a1
not modelled
5.5
22
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Nitrogenase iron protein-like43 d1u9pa1
not modelled
5.5
29
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: Arc/Mnt-like phage repressors44 c2w45A_
not modelled
5.4
18
PDB header: hydrolaseChain: A: PDB Molecule: alkaline exonuclease;PDBTitle: epstein-barr virus alkaline nuclease
45 d2p0ma1
not modelled
5.4
12
Fold: LipoxigenaseSuperfamily: LipoxigenaseFamily: Animal lipoxigenases46 c2dmpA_
not modelled
5.3
30
PDB header: dna binding proteinChain: A: PDB Molecule: zinc fingers and homeoboxes protein 2;PDBTitle: solution structure of the third homeobox domain of zinc2 fingers and homeoboxes protein 2
47 d1ydha_
not modelled
5.1
10
Fold: MCP/YpsA-likeSuperfamily: MCP/YpsA-likeFamily: MoCo carrier protein-like48 d1upka_
not modelled
5.1
14
Fold: alpha-alpha superhelixSuperfamily: ARM repeatFamily: Mo25 protein