| 1 |
|
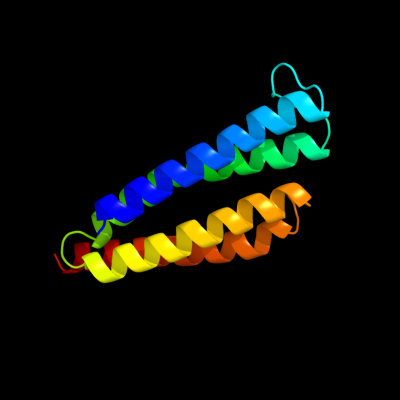
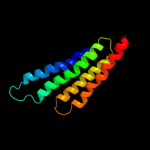
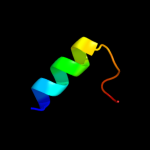
PDB 2pno chain L
Region: 2 - 125
Aligned: 122
Modelled: 124
Confidence: 100.0%
Identity: 25%
PDB header:lyase
Chain: L: PDB Molecule:leukotriene c4 synthase;
PDBTitle: crystal structure of human leukotriene c4 synthase
Phyre2
| 2 |
|
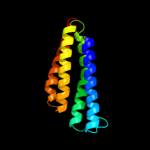
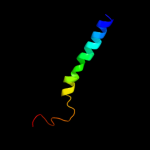
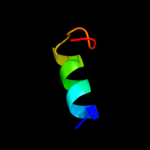
PDB 2uui chain A domain 1
Region: 2 - 125
Aligned: 122
Modelled: 124
Confidence: 100.0%
Identity: 25%
Fold: MAPEG domain-like
Superfamily: MAPEG domain-like
Family: MAPEG domain
Phyre2
| 3 |
|
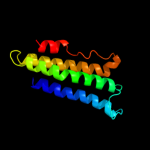
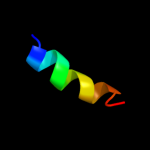
PDB 2q7r chain A domain 1
Region: 3 - 120
Aligned: 116
Modelled: 114
Confidence: 100.0%
Identity: 13%
Fold: MAPEG domain-like
Superfamily: MAPEG domain-like
Family: MAPEG domain
Phyre2
| 4 |
|
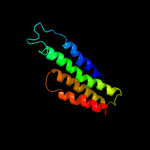
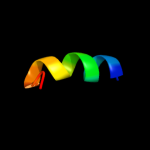
PDB 2h8a chain A domain 1
Region: 2 - 116
Aligned: 113
Modelled: 115
Confidence: 100.0%
Identity: 18%
Fold: MAPEG domain-like
Superfamily: MAPEG domain-like
Family: MAPEG domain
Phyre2
| 5 |
|
PDB 3dww chain A
Region: 2 - 126
Aligned: 125
Modelled: 125
Confidence: 100.0%
Identity: 18%
PDB header:isomerase
Chain: A: PDB Molecule:prostaglandin e synthase;
PDBTitle: electron crystallographic structure of human microsomal2 prostaglandin e synthase 1
Phyre2
| 6 |
|
PDB 2knc chain A
Region: 2 - 39
Aligned: 38
Modelled: 38
Confidence: 11.4%
Identity: 13%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 7 |
|
PDB 1znf chain A
Region: 36 - 49
Aligned: 14
Modelled: 14
Confidence: 6.5%
Identity: 36%
PDB header:zinc finger dna binding domain
Chain: A: PDB Molecule:31st zinc finger from xfin;
PDBTitle: three-dimensional solution structure of a single zinc2 finger dna-binding domain
Phyre2
| 8 |
|
PDB 1znf chain A
Region: 36 - 49
Aligned: 14
Modelled: 14
Confidence: 6.5%
Identity: 36%
Fold: beta-beta-alpha zinc fingers
Superfamily: beta-beta-alpha zinc fingers
Family: Classic zinc finger, C2H2
Phyre2
| 9 |
|
PDB 3pmd chain A
Region: 17 - 34
Aligned: 18
Modelled: 18
Confidence: 6.2%
Identity: 28%
PDB header:lipid binding protein
Chain: A: PDB Molecule:conserved domain protein;
PDBTitle: crystal structure of the sporulation inhibitor pxo1-118 from bacillus2 anthracis
Phyre2
| 10 |
|
PDB 1yku chain B
Region: 17 - 34
Aligned: 18
Modelled: 18
Confidence: 6.0%
Identity: 22%
PDB header:unknown function
Chain: B: PDB Molecule:hypothetical protein pxo2-61;
PDBTitle: crystal structure of a sensor domain homolog
Phyre2