| 1 |
|
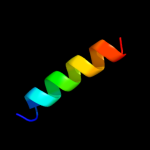
PDB 1b9u chain A
Region: 17 - 34
Aligned: 18
Modelled: 18
Confidence: 40.2%
Identity: 56%
PDB header:hydrolase
Chain: A: PDB Molecule:protein (atp synthase);
PDBTitle: membrane domain of the subunit b of the e.coli atp synthase
Phyre2
| 2 |
|
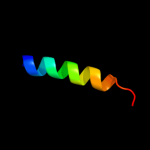
PDB 1q90 chain N
Region: 1 - 10
Aligned: 10
Modelled: 10
Confidence: 13.3%
Identity: 40%
Fold: Single transmembrane helix
Superfamily: PetN subunit of the cytochrome b6f complex
Family: PetN subunit of the cytochrome b6f complex
Phyre2
| 3 |
|
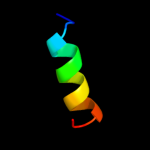
PDB 3mq1 chain A
Region: 42 - 60
Aligned: 19
Modelled: 19
Confidence: 10.5%
Identity: 32%
PDB header:allergen
Chain: A: PDB Molecule:mite allergen der p 5;
PDBTitle: crystal structure of dust mite allergen der p 5
Phyre2
| 4 |
|
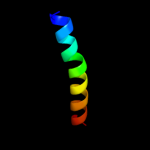
PDB 2jmh chain A
Region: 42 - 60
Aligned: 19
Modelled: 19
Confidence: 9.5%
Identity: 21%
PDB header:allergen
Chain: A: PDB Molecule:mite allergen blo t 5;
PDBTitle: nmr solution structure of blo t 5, a major mite allergen2 from blomia tropicalis
Phyre2
| 5 |
|
PDB 2i7n chain A domain 2
Region: 69 - 85
Aligned: 17
Modelled: 17
Confidence: 7.7%
Identity: 53%
Fold: Ribonuclease H-like motif
Superfamily: Actin-like ATPase domain
Family: Fumble-like
Phyre2
| 6 |
|
PDB 2axt chain M domain 1
Region: 23 - 63
Aligned: 26
Modelled: 26
Confidence: 5.6%
Identity: 50%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction center protein M, PsbM
Family: PsbM-like
Phyre2
| 7 |
|
PDB 2roc chain B
Region: 38 - 56
Aligned: 19
Modelled: 19
Confidence: 5.4%
Identity: 42%
PDB header:apoptosis
Chain: B: PDB Molecule:bcl-2-binding component 3;
PDBTitle: solution structure of mcl-1 complexed with puma
Phyre2