| 1 |
|
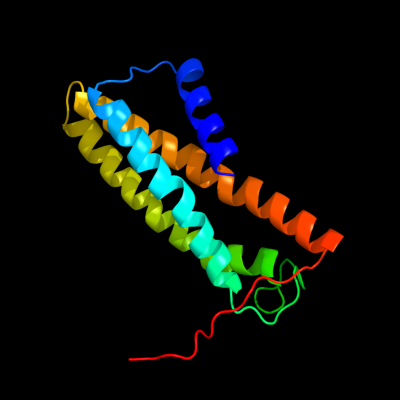
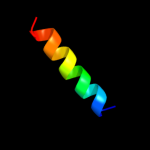
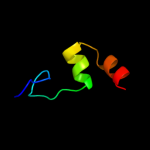
PDB 3aqp chain B
Region: 220 - 364
Aligned: 136
Modelled: 145
Confidence: 23.3%
Identity: 15%
PDB header:membrane protein
Chain: B: PDB Molecule:probable secdf protein-export membrane protein;
PDBTitle: crystal structure of secdf, a translocon-associated membrane protein,2 from thermus thrmophilus
Phyre2
| 2 |
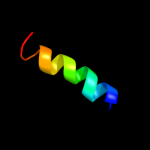
|
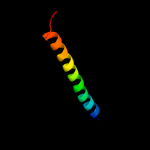
PDB 3k07 chain A
Region: 184 - 357
Aligned: 167
Modelled: 174
Confidence: 12.3%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:cation efflux system protein cusa;
PDBTitle: crystal structure of cusa
Phyre2
| 3 |
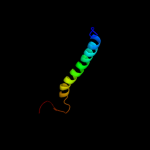
|
PDB 3izc chain Q
Region: 345 - 362
Aligned: 18
Modelled: 18
Confidence: 9.5%
Identity: 33%
PDB header:ribosome
Chain: Q: PDB Molecule:60s ribosomal protein rpl5 (l18p);
PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
Phyre2
| 4 |
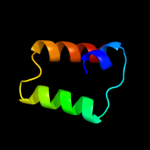
|
PDB 3u5i chain N
Region: 345 - 362
Aligned: 18
Modelled: 18
Confidence: 9.5%
Identity: 33%
PDB header:ribosome
Chain: N: PDB Molecule:60s ribosomal protein l15-a;
PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution
Phyre2
| 5 |
|
PDB 3u5e chain N
Region: 345 - 362
Aligned: 18
Modelled: 18
Confidence: 9.5%
Identity: 33%
PDB header:ribosome
Chain: N: PDB Molecule:60s ribosomal protein l15-a;
PDBTitle: the structure of the eukaryotic ribosome at 3.0 resolution
Phyre2
| 6 |
|
PDB 3izs chain Q
Region: 345 - 362
Aligned: 18
Modelled: 18
Confidence: 9.5%
Identity: 33%
PDB header:ribosome
Chain: Q: PDB Molecule:60s ribosomal protein rpl5 (l18p);
PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
Phyre2
| 7 |
|
PDB 2knc chain A
Region: 315 - 362
Aligned: 48
Modelled: 48
Confidence: 7.9%
Identity: 23%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 8 |
|
PDB 1uou chain A domain 1
Region: 262 - 303
Aligned: 42
Modelled: 42
Confidence: 6.1%
Identity: 12%
Fold: Methionine synthase domain-like
Superfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
Family: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain
Phyre2
| 9 |
|
PDB 2cpw chain A domain 1
Region: 265 - 299
Aligned: 34
Modelled: 35
Confidence: 5.7%
Identity: 15%
Fold: RuvA C-terminal domain-like
Superfamily: UBA-like
Family: UBA domain
Phyre2
| 10 |
|
PDB 2rmz chain A
Region: 316 - 353
Aligned: 38
Modelled: 38
Confidence: 5.2%
Identity: 24%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin beta-3;
PDBTitle: bicelle-embedded integrin beta3 transmembrane segment
Phyre2
| 11 |
|
PDB 3iz5 chain Q
Region: 345 - 362
Aligned: 18
Modelled: 18
Confidence: 5.1%
Identity: 33%
PDB header:ribosome
Chain: Q: PDB Molecule:60s ribosomal protein l5 (l18p);
PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
Phyre2