| 1 |
|
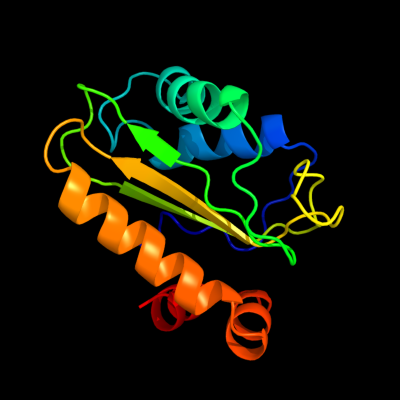
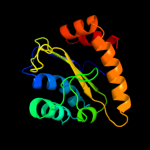
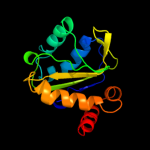
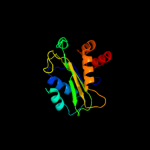
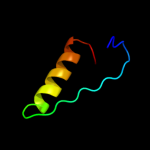
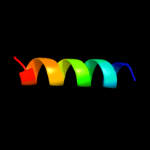
PDB 2a0j chain A
Region: 3 - 143
Aligned: 140
Modelled: 141
Confidence: 100.0%
Identity: 27%
PDB header:transferase
Chain: A: PDB Molecule:pts system, nitrogen regulatory iia protein;
PDBTitle: crystal structure of nitrogen regulatory protein iia-ntr from2 neisseria meningitidis
Phyre2
| 2 |
|
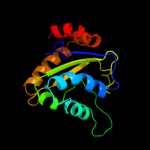
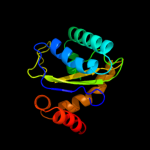
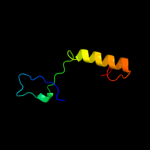
PDB 3urr chain B
Region: 3 - 145
Aligned: 140
Modelled: 143
Confidence: 100.0%
Identity: 26%
PDB header:transferase
Chain: B: PDB Molecule:pts iia-like nitrogen-regulatory protein ptsn;
PDBTitle: structure of pts iia-like nitrogen-regulatory protein ptsn (bth_i0484)2 (ptsn)
Phyre2
| 3 |
|
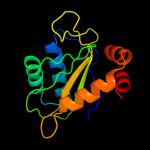
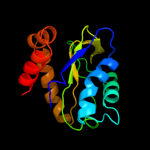
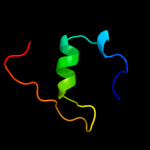
PDB 1a6j chain A
Region: 1 - 145
Aligned: 144
Modelled: 145
Confidence: 100.0%
Identity: 21%
Fold: Phoshotransferase/anion transport protein
Superfamily: Phoshotransferase/anion transport protein
Family: IIA domain of mannitol-specific and ntr phosphotransferase EII
Phyre2
| 4 |
|
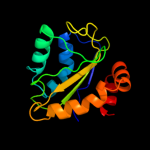
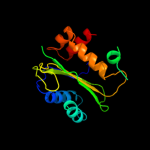
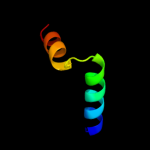
PDB 3oxp chain B
Region: 3 - 145
Aligned: 139
Modelled: 143
Confidence: 100.0%
Identity: 16%
PDB header:transferase
Chain: B: PDB Molecule:phosphotransferase enzyme ii, a component;
PDBTitle: structure of phosphotransferase enzyme ii, a component from yersinia2 pestis co92 at 1.2 a resolution
Phyre2
| 5 |
|
PDB 3oxp chain A
Region: 3 - 145
Aligned: 139
Modelled: 143
Confidence: 100.0%
Identity: 16%
PDB header:transferase
Chain: A: PDB Molecule:phosphotransferase enzyme ii, a component;
PDBTitle: structure of phosphotransferase enzyme ii, a component from yersinia2 pestis co92 at 1.2 a resolution
Phyre2
| 6 |
|
PDB 1a3a chain A
Region: 1 - 143
Aligned: 138
Modelled: 143
Confidence: 100.0%
Identity: 23%
Fold: Phoshotransferase/anion transport protein
Superfamily: Phoshotransferase/anion transport protein
Family: IIA domain of mannitol-specific and ntr phosphotransferase EII
Phyre2
| 7 |
|
PDB 2oq3 chain A
Region: 2 - 144
Aligned: 138
Modelled: 143
Confidence: 100.0%
Identity: 16%
PDB header:transferase
Chain: A: PDB Molecule:mannitol-specific cryptic phosphotransferase
PDBTitle: solution structure of the mannitol- specific cryptic2 phosphotransferase enzyme iia cmtb from escherichia coli
Phyre2
| 8 |
|
PDB 3bjv chain A
Region: 2 - 148
Aligned: 142
Modelled: 145
Confidence: 100.0%
Identity: 18%
PDB header:transferase
Chain: A: PDB Molecule:rmpa;
PDBTitle: the crystal structure of a putative pts iia(ptxa) from streptococcus2 mutans
Phyre2
| 9 |
|
PDB 2oqt chain D
Region: 3 - 148
Aligned: 141
Modelled: 144
Confidence: 100.0%
Identity: 13%
PDB header:transferase
Chain: D: PDB Molecule:hypothetical protein spy0176;
PDBTitle: structural genomics, the crystal structure of a putative2 pts iia domain from streptococcus pyogenes m1 gas
Phyre2
| 10 |
|
PDB 1xiz chain A
Region: 3 - 147
Aligned: 143
Modelled: 145
Confidence: 100.0%
Identity: 20%
Fold: Phoshotransferase/anion transport protein
Superfamily: Phoshotransferase/anion transport protein
Family: IIA domain of mannitol-specific and ntr phosphotransferase EII
Phyre2
| 11 |
|
PDB 1hyn chain Q
Region: 4 - 146
Aligned: 143
Modelled: 143
Confidence: 98.3%
Identity: 20%
PDB header:membrane protein
Chain: Q: PDB Molecule:band 3 anion transport protein;
PDBTitle: crystal structure of the cytoplasmic domain of human2 erythrocyte band-3 protein
Phyre2
| 12 |
|
PDB 1hyn chain P
Region: 4 - 146
Aligned: 143
Modelled: 143
Confidence: 98.3%
Identity: 20%
Fold: Phoshotransferase/anion transport protein
Superfamily: Phoshotransferase/anion transport protein
Family: Anion transport protein, cytoplasmic domain
Phyre2
| 13 |
|
PDB 2v8k chain A
Region: 81 - 129
Aligned: 44
Modelled: 49
Confidence: 25.2%
Identity: 14%
PDB header:lyase
Chain: A: PDB Molecule:pectate lyase;
PDBTitle: structure of a family 2 pectate lyase in complex with2 trigalacturonic acid
Phyre2
| 14 |
|
PDB 1gjj chain A domain 1
Region: 33 - 67
Aligned: 30
Modelled: 35
Confidence: 9.8%
Identity: 23%
Fold: LEM/SAP HeH motif
Superfamily: LEM domain
Family: LEM domain
Phyre2
| 15 |
|
PDB 3dfg chain A
Region: 19 - 47
Aligned: 28
Modelled: 29
Confidence: 6.1%
Identity: 11%
PDB header:recombination
Chain: A: PDB Molecule:regulatory protein recx;
PDBTitle: crystal structure of recx: a potent inhibitor protein of2 reca from xanthomonas campestris
Phyre2
| 16 |
|
PDB 1pch chain A
Region: 1 - 36
Aligned: 36
Modelled: 36
Confidence: 6.1%
Identity: 22%
Fold: HPr-like
Superfamily: HPr-like
Family: HPr-like
Phyre2
| 17 |
|
PDB 2voi chain B
Region: 17 - 33
Aligned: 17
Modelled: 17
Confidence: 6.0%
Identity: 18%
PDB header:apoptosis
Chain: B: PDB Molecule:bh3-interacting domain death agonist p13;
PDBTitle: structure of mouse a1 bound to the bid bh3-domain
Phyre2