




Progress Protein In Structure Assignment
Structure Assignenment Summer 1998 & Summer 1999
The pie-charts show the progress in structural assignment for the genome of
Mycoplasma genitalium. A subset of the Mycoplasma genitalium
genome that was structurally completely unassigned in early summer 1999
takining into account the results of several research groups (see,
Teichmann SA, Chothia C, Gerstein M (1999), Curr. Opin. Struc. Biol. 9: 390-399. and , http://www.mrc-lmb.cam.ac.uk:80/genomes/MG/).
We analysed this subset of the genome with PSI-BLAST and the most recent
databases (sequence and structure). Two percentages of the residues of this
subset could be assigned via close homology to sequence of known structure and
6 percentages to proteins of known structure via remote homology only. That
means in about 1 years time there was a growth in structure assignment of 8
percentages. This is not only due to the growth of the structure database but
also dependent on the growth of the protein sequence database because
PSI-BLAST needs intermediate sequences to find remote homologies.
In an others years time one can expect more then 50% of the residues in the
genome to be structurally annotated. Taking into account a much more rapid
growth of the databases the assigned fraction will probably much larger.
The pie-charts below show the results of our structure assignment
with databases from 1998 (left) and the 1999 (right). Fraction are calculated
based on residues.
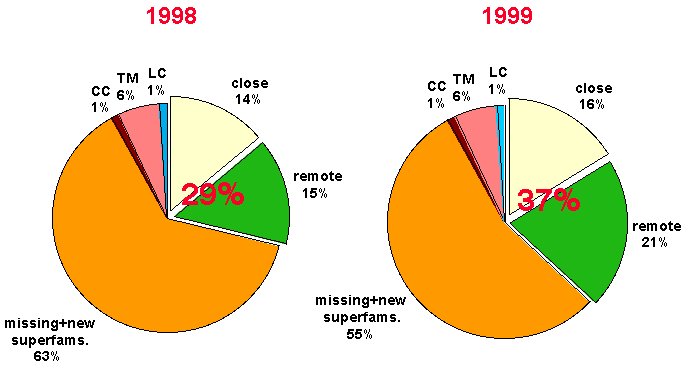
Legend:
in CC (coiled-coils),
TM (Transmembrane helices),
LC (low complexety regions),
close (matches by close homologues),
remote (matches remote homologues only),
missing+new superfams (undetected homologues
and new structural superfamilies)
get structural annotations for Mycoplasma genitalium ...





Copyright © 1999-2002 Cancer Research UK
All Rights Reserved, disclaimer
Comments to author: a.mueller@cancer.org.uk
Generated: Thu Jun 27, 2002

