





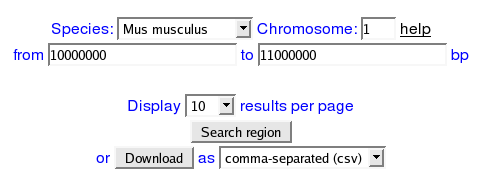
HOW CAN ONE SEARCH THE DATABASE BY REGION?
By choosing which species from the dropdown menu and submitting the chromosome number, start and end base pairs of the region the user is interested in, a list with all the
SNPSTRs in the area is obtained.
e.g. I want to get all SNPSTRs that exist in mouse chromosome 1 from base pair 100000000 to base pair 11000000.
I will input the following values:
WHY IS THERE A SEARCH AND A DOWNLOAD BUTTON?
By default the results are displayed as HTML which is great when one is looking for a small number of SNPSTRs. However when more than a few SNPSTRs are being looked at, the HTML
format is not very useful. In addition, one might want to get the information in a format which can be easily manipulated for example, one my want to input the data into a Perl
Script for further research, something that HTML output would not allow.
For this reason, the database interface provides the option of downloading the data rather than viewing
them. The user can choose if he or she wants the file to be tab-delimited or comma-delimited, and when he or she clicks on the download button a window pops up that asks if the
user wants to open the file or save it in their computer. The files contain all the information contained in the HTML page with the exception of the multiple gene accession ids
(only the Ensembl gene ids are provided).




