





WHAT IS A SNPSTR?
SNPSTRs are a relatively new type of compound genetic marker which combines a STR marker with one or more tightly linked SNPs. This combination of
co-inherited markers evolving at different rates may offer the possibility of gaining better resolved insights into population genetic processes
compared to when these different marker types are used separately. SNPSTRs were first described by Mountain et al (2002) who
developed experimental protocols for autosomal SNPSTRs which contain a SNP and a microsatellite within 500 base pairs apart.
Here, the SNP(s) and the microsatellite are less than 250 base pairs apart so a SNPSTR could look like this:
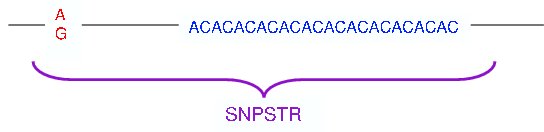 Because of the proximity of the microsatellite to the SNP, SNPSTRs have the advantage that:
Because of the proximity of the microsatellite to the SNP, SNPSTRs have the advantage that:
- they are not broken up by recombination;
- significant difference in mutation rate between polymorphisms so that they can provide complementary evolutionary information and
- they contain slowly evolving binary markers (the SNP) as well as the quickly evolving microsatellites
In principle at least, it should therefore be possible to infer the age of the SNP allele (or the most recent common ancestor of all individuals
carrying that allele) from the microsatellite data (using a generic model of the microsatellite mutation process). Each SNPSTR acts as a "mini Y-chromosome"
and combining many unlinked SNPSTRs will give us a rich data-source to infer past demographic events (or test for deviations from a neutral model).
HOW WERE SNPSTRS IDENTIFIED?
To identify SNPSTRs we started with SNPSTR sequence identification and then used the genomic positions of SNPs to identify nearby genes and disease regions and to obtain genetic variation
information as can be seen in the following flow chart.
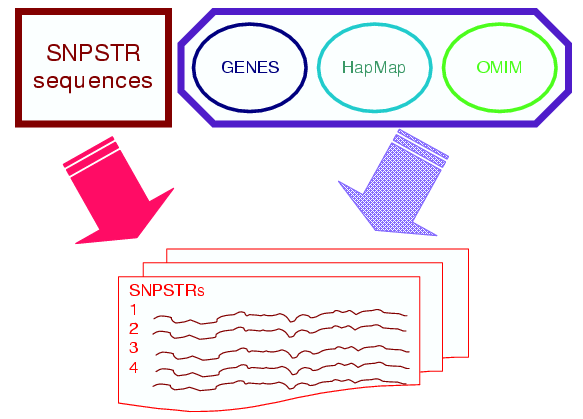
SNPSTRs were identified for five model species, those where SNP information existed in both of the NCBI and
Ensembl databases. These species are human (Homo sapiens), mouse (Mus musculus), rat (Rattus norvegicus), dog (Canis
familiaris), and chicken (Gallus gallus).
The means chosen to extract the sequences was the Ensembl Perl Application Programming Interface (API) which is an interface
written in object-oriented Perl that allows external users to extract particular data, to customise Ensembl or to store additional data in the database. A 1001 base pair long sequence was
retrieved for each SNP that contains the SNP exactly in the middle, satisfying thus the requirement above. These sequences were scanned for microsatellites with
Tandem Repeats Finder (TRF) which locates and displays tandem repeats in DNA sequences by moving a sliding window along the sequences to seek candidate matched adjacent repeats of any size in
DNA, including repeats containing mismatches and indels.
Variation information was obtained for human SNPSTRs in the form of allele counts from the ftp site of the HapMap project database. The aim was to use this
information not only to find the polymorphism levels of the SNPs in the different populations in terms of heterozygosity but also to calculate FST values to identify those SNPs that
are differently polymorphic in the different populations.
The second source of extra information obtained was the positions of coding genes in the human genome. These were used to identify which SNPSTRs were in genes (exons and introns) or in intergenic
sequences. If genes are more affected by natural selection you would expect those SNPSTRs in or near genes to be more conserved than the others. The SNP part of the SNPSTR should be less
polymorphic and the length of the microsatellite part should be less variable. Gene and exon coordinates were obtained using the Ensembl API. Finally, disease information was obtained to identify those SNPSTRs that were found in disease areas. The Mendelian Inheritance in Man project is a database that catalogues all the known diseases
with a genetic component, and - when possible - links them to the relevant genes in the human genome. OMIM gene coordinates were obtained using the Ensembl API.
WHAT DOES EACH SNPSTR ENTRY CONTAIN?
For each SNPSTR the following information is contained in the database:
- SNPSTR database id
- species
- chromosome number
- genomic start and end coordinates
- microsatellite start and end coordinates
- microsatellite repeat unit length, sequence and copy number
- information on whether the microsatellite consists only of perfect repeats or if it contains some non-perfect repeats
- SNP genomic location, allele counts for the four populations, HS and FST values
- accesion numbers of the nearest gene from Ensembl, uniprot, Entrez Gene and HUGO databases as well as Pubmed ID for paper that mentions the gene
- name of nearest OMIM disease where applicable and the distance of the SNP compared to that
- SNPSTR sequence
WHAT ARE THE DATABASE CONTENTS (Release 1.4)?
In the SNPSTR database we catalogue all inferable SNPSTRs for the five model species, where sufficient SNP information exists in both of the NCBI and Ensembl databases.
These species are human (Homo sapiens), mouse (Mus musculus), rat (Rattus norvegicus), dog (Canis familiaris) and chicken (Gallus gallus).
Release 1.5 of the SNPSTR database (Mar 2008) contains:
| SPECIES | SNPSTR | GENIC | EXONIC | INTRONIC |
| HUMAN | 852,777 | 285,081 | 8,221 | 276,860 |
| MOUSE | 1,116,643 | 396,183 | 12,041 | 384,142 |
| RAT | 1,539 | 905 | 558 | 347 |
| DOG | 16,886 | 3,438 | 106 | 3,332 |
| CHICKEN | 39,848 | 11,850 | 398 | 11,452 |
| TOTAL | 2,027,693 | 697,457 | 21,324 | 676,133 |
Previous Releases
I USED THE DATABASE. HOW DO I CITE IT?
Please cite:
Agrafioti I, Stumpf MP (2007) SNPSTR: a database of compound microsatellite-SNP markers. Nucleic Acids Res. 35(Database issue):D71-5





